Origin of land plants: do conjugating green algae hold the key?
- PMID: 21501468
- PMCID: PMC3088898
- DOI: 10.1186/1471-2148-11-104
Origin of land plants: do conjugating green algae hold the key?
Abstract
Background: The terrestrial habitat was colonized by the ancestors of modern land plants about 500 to 470 million years ago. Today it is widely accepted that land plants (embryophytes) evolved from streptophyte algae, also referred to as charophycean algae. The streptophyte algae are a paraphyletic group of green algae, ranging from unicellular flagellates to morphologically complex forms such as the stoneworts (Charales). For a better understanding of the evolution of land plants, it is of prime importance to identify the streptophyte algae that are the sister-group to the embryophytes. The Charales, the Coleochaetales or more recently the Zygnematales have been considered to be the sister group of the embryophytes However, despite many years of phylogenetic studies, this question has not been resolved and remains controversial.
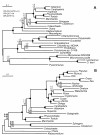
Results: Here, we use a large data set of nuclear-encoded genes (129 proteins) from 40 green plant taxa (Viridiplantae) including 21 embryophytes and six streptophyte algae, representing all major streptophyte algal lineages, to investigate the phylogenetic relationships of streptophyte algae and embryophytes. Our phylogenetic analyses indicate that either the Zygnematales or a clade consisting of the Zygnematales and the Coleochaetales are the sister group to embryophytes.
Conclusions: Our analyses support the notion that the Charales are not the closest living relatives of embryophytes. Instead, the Zygnematales or a clade consisting of Zygnematales and Coleochaetales are most likely the sister group of embryophytes. Although this result is in agreement with a previously published phylogenetic study of chloroplast genomes, additional data are needed to confirm this conclusion. A Zygnematales/embryophyte sister group relationship has important implications for early land plant evolution. If substantiated, it should allow us to address important questions regarding the primary adaptations of viridiplants during the conquest of land. Clearly, the biology of the Zygnematales will receive renewed interest in the future.
© 2011 Wodniok et al; licensee BioMed Central Ltd.
Figures

References
-
- Lang D, Weiche B, Timmerhaus G, Richardt S, Riano-Pachon DM, Correa LGG, Reski R, Mueller-Roeber B, Rensing SA. Genome-Wide Phylogenetic Comparative Analysis of Plant Transcriptional Regulation: A Timeline of Loss, Gain, Expansion, and Correlation with Complexity. Genome Biol Evol. pp. 488–503. - DOI - PMC - PubMed
-
- Graham LE. Origin of land plants. New York: John Wiley & Sons, Inc.; 1993.
-
- Kenrick P, Crane PR. The origin and early diversification of land plants. Washington, London: Smithsonian Institution Press; 1997.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

