Thermostabilisation of an agonist-bound conformation of the human adenosine A(2A) receptor
- PMID: 21501622
- PMCID: PMC3145977
- DOI: 10.1016/j.jmb.2011.03.075
Thermostabilisation of an agonist-bound conformation of the human adenosine A(2A) receptor
Abstract
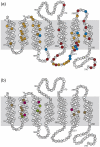
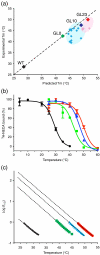
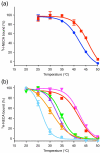
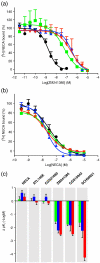
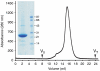
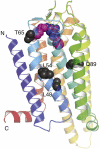
The adenosine A(2A) receptor (A(2A)R) is a G-protein-coupled receptor that plays a key role in transmembrane signalling mediated by the agonist adenosine. The structure of A(2A)R was determined recently in an antagonist-bound conformation, which was facilitated by the T4 lysozyme fusion in cytoplasmic loop 3 and the considerable stabilisation conferred on the receptor by the bound inverse agonist ZM241385. Unfortunately, the natural agonist adenosine does not sufficiently stabilise the receptor for the formation of diffraction-quality crystals. As a first step towards determining the structure of A(2A)R bound to an agonist, the receptor was thermostabilised by systematic mutagenesis in the presence of the bound agonist [(3)H]5'-N-ethylcarboxamidoadenosine (NECA). Four thermostabilising mutations were identified that when combined to give mutant A(2A)R-GL26, conferred a greater than 200-fold decrease in its rate of unfolding compared to the wild-type receptor. Pharmacological analysis suggested that A(2A)R-GL26 is stabilised in an agonist-bound conformation because antagonists bind with up to 320-fold decreased affinity. None of the thermostabilising mutations are in the ZM241385 binding pocket, suggesting that the mutations affect ligand binding by altering the conformation of the receptor rather than through direct interactions with ligands. A(2A)R-GL26 shows considerable stability in short-chain detergents, which has allowed its purification and crystallisation.
Crown Copyright © 2011. Published by Elsevier Ltd. All rights reserved.
Figures
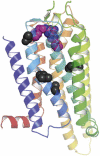
References
-
- Kobilka B.K., Deupi X. Conformational complexity of G-protein-coupled receptors. Trends Pharmacol. Sci. 2007;28:397–406. - PubMed
-
- DeWire S.M., Ahn S., Lefkowitz R.J., Shenoy S.K. Beta-arrestins and cell signaling. Annu. Rev. Physiol. 2007;69:483–510. - PubMed
-
- Hopkins A.L., Groom C.R. The druggable genome. Nat. Rev., Drug Discov. 2002;1:727–730. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

