Mitochondrial dysfunction impairs tumor suppressor p53 expression/function
- PMID: 21502317
- PMCID: PMC3121470
- DOI: 10.1074/jbc.M110.163063
Mitochondrial dysfunction impairs tumor suppressor p53 expression/function
Abstract
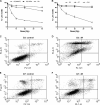
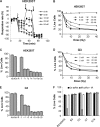
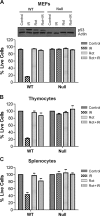
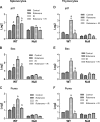
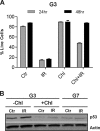
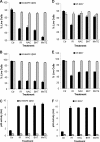
Recently, mitochondria have been suggested to act in tumor suppression. However, the underlying mechanisms by which mitochondria suppress tumorigenesis are far from being clear. In this study, we have investigated the link between mitochondrial dysfunction and the tumor suppressor protein p53 using a set of respiration-deficient (Res(-)) mammalian cell mutants with impaired assembly of the oxidative phosphorylation machinery. Our data suggest that normal mitochondrial function is required for γ-irradiation (γIR)-induced cell death, which is mainly a p53-dependent process. The Res(-) cells are protected against γIR-induced cell death due to impaired p53 expression/function. We find that the loss of complex I biogenesis in the absence of the MWFE subunit reduces the steady-state level of the p53 protein, although there is no effect on the p53 protein level in the absence of the ESSS subunit that is also essential for complex I assembly. The p53 protein level was also reduced to undetectable levels in Res(-) cells with severely impaired mitochondrial protein synthesis. This suggests that p53 protein expression is differentially regulated depending upon the type of electron transport chain/respiratory chain deficiency. Moreover, irrespective of the differences in the p53 protein expression profile, γIR-induced p53 activity is compromised in all Res(-) cells. Using two different conditional systems for complex I assembly, we also show that the effect of mitochondrial dysfunction on p53 expression/function is a reversible phenomenon. We believe that these findings will have major implications in the understanding of cancer development and therapy.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

