Histone H4 acetylation differentially modulates arginine methylation by an in Cis mechanism
- PMID: 21502321
- PMCID: PMC3121530
- DOI: 10.1074/jbc.M110.207258
Histone H4 acetylation differentially modulates arginine methylation by an in Cis mechanism
Abstract
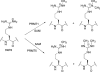
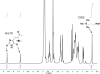
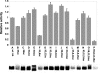
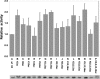
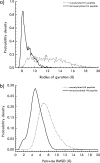
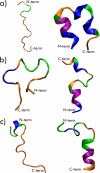
Histone H4 undergoes extensive post-translational modifications (PTMs) at its N-terminal tail. Many of these PTMs profoundly affect the on and off status of gene transcription. The molecular mechanism by which histone PTMs modulate genetic and epigenetic processes is not fully understood. In particular, how a PTM mark affects the presence and level of other histone modification marks needs to be addressed and is essential for better understanding the molecular basis of histone code hypothesis. To dissect the interplaying relationship between different histone modification marks, we investigated how individual lysine acetylations and their different combinations at the H4 tail affect Arg-3 methylation in cis. Our data reveal that the effect of lysine acetylation on arginine methylation depends on the site of acetylation and the type of methylation. Although certain acetylations present a repressive impact on PRMT1-mediated methylation (type I methylation), lysine acetylation generally is correlated with enhanced methylation by PRMT5 (type II dimethylation). In particular, Lys-5 acetylation decreases the activity of PRMT1 but increases that of PRMT5. Furthermore, circular dichroism study and computer simulation demonstrate that hyperacetylation increases the content of ordered secondary structures at the H4 tail region. These findings provide new insights into the regulatory mechanism of Arg-3 methylation by H4 acetylation and unravel the complex intercommunications that exist between different the PTM marks in cis. The divergent activities of PRMT1 and PRMT5 with respect to different acetyl-H4 substrates suggest that type I and type II protein-arginine methyltransferases use distinct molecular determinants for substrate recognition and catalysis.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

