A 3D analysis of yeast ER structure reveals how ER domains are organized by membrane curvature
- PMID: 21502358
- PMCID: PMC3080256
- DOI: 10.1083/jcb.201011039
A 3D analysis of yeast ER structure reveals how ER domains are organized by membrane curvature
Abstract
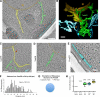
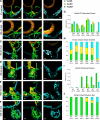
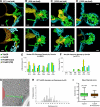
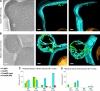
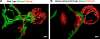
We analyzed the structure of yeast endoplasmic reticulum (ER) during six sequential stages of budding by electron tomography to reveal a three-dimensional portrait of ER organization during inheritance at a nanometer resolution. We have determined the distribution, dimensions, and ribosome densities of structurally distinct but continuous ER domains during multiple stages of budding with and without the tubule-shaping proteins, reticulons (Rtns) and Yop1. In wild-type cells, the peripheral ER contains cytoplasmic cisternae, many tubules, and a large plasma membrane (PM)-associated ER domain that consists of both tubules and fenestrated cisternae. In the absence of Rtn/Yop1, all three domains lose membrane curvature, ER ribosome density changes, and the amount of PM-associated ER increases dramatically. Deletion of Rtns/Yop1 does not, however, prevent bloated ER tubules from being pulled from the mother cisterna into the bud and strongly suggests that Rtns/Yop1 stabilize/maintain rather than generate membrane curvature at all peripheral ER domains in yeast.
Figures


References
-
- Achleitner G., Gaigg B., Krasser A., Kainersdorfer E., Kohlwein S.D., Perktold A., Zellnig G., Daum G. 1999. Association between the endoplasmic reticulum and mitochondria of yeast facilitates interorganelle transport of phospholipids through membrane contact. Eur. J. Biochem. 264:545–553 10.1046/j.1432-1327.1999.00658.x - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

