Aprataxin localizes to mitochondria and preserves mitochondrial function
- PMID: 21502511
- PMCID: PMC3088601
- DOI: 10.1073/pnas.1100084108
Aprataxin localizes to mitochondria and preserves mitochondrial function
Abstract
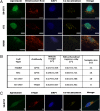
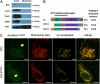
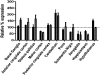
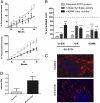
Ataxia with oculomotor apraxia 1 is caused by mutation in the APTX gene, which encodes the DNA strand-break repair protein aprataxin. Aprataxin exhibits homology to the histidine triad superfamily of nucleotide hydrolases and transferases and removes 5'-adenylate groups from DNA that arise from aborted ligation reactions. We report herein that aprataxin localizes to mitochondria in human cells and we identify an N-terminal amino acid sequence that targets certain isoforms of the protein to this intracellular compartment. We also show that transcripts encoding this unique N-terminal stretch are expressed in the human brain, with highest production in the cerebellum. Depletion of aprataxin in human SH-SY5Y neuroblastoma cells and primary skeletal muscle myoblasts results in mitochondrial dysfunction, which is revealed by reduced citrate synthase activity and mtDNA copy number. Moreover, mtDNA, not nuclear DNA, was found to have higher levels of background DNA damage on aprataxin knockdown, suggesting a direct role for the enzyme in mtDNA processing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates.Nature. 2006 Oct 12;443(7112):713-6. doi: 10.1038/nature05164. Epub 2006 Sep 10. Nature. 2006. PMID: 16964241
-
Actions of aprataxin in multiple DNA repair pathways.J Biol Chem. 2007 Mar 30;282(13):9469-9474. doi: 10.1074/jbc.M611489200. Epub 2007 Feb 2. J Biol Chem. 2007. PMID: 17276982
-
Aprataxin forms a discrete branch in the HIT (histidine triad) superfamily of proteins with both DNA/RNA binding and nucleotide hydrolase activities.J Biol Chem. 2006 May 19;281(20):13939-48. doi: 10.1074/jbc.M507946200. Epub 2006 Mar 16. J Biol Chem. 2006. PMID: 16547001
-
The role of TDP1 and APTX in mitochondrial DNA repair.Biochimie. 2014 May;100:121-4. doi: 10.1016/j.biochi.2013.10.011. Epub 2013 Oct 22. Biochimie. 2014. PMID: 24161509 Free PMC article. Review.
-
Molecular underpinnings of Aprataxin RNA/DNA deadenylase function and dysfunction in neurological disease.Prog Biophys Mol Biol. 2015 Mar;117(2-3):157-165. doi: 10.1016/j.pbiomolbio.2015.01.007. Epub 2015 Jan 29. Prog Biophys Mol Biol. 2015. PMID: 25637650 Free PMC article. Review.
Cited by
-
Mitochondrial Genetic and Epigenetic Regulations in Cancer: Therapeutic Potential.Int J Mol Sci. 2022 Jul 18;23(14):7897. doi: 10.3390/ijms23147897. Int J Mol Sci. 2022. PMID: 35887244 Free PMC article. Review.
-
Mitochondrial determinants of cancer health disparities.Semin Cancer Biol. 2017 Dec;47:125-146. doi: 10.1016/j.semcancer.2017.05.001. Epub 2017 May 6. Semin Cancer Biol. 2017. PMID: 28487205 Free PMC article. Review.
-
Slow mitochondrial repair of 5'-AMP renders mtDNA susceptible to damage in APTX deficient cells.Sci Rep. 2015 Aug 10;5:12876. doi: 10.1038/srep12876. Sci Rep. 2015. PMID: 26256098 Free PMC article.
-
Complementation of aprataxin deficiency by base excision repair enzymes in mitochondrial extracts.Nucleic Acids Res. 2017 Sep 29;45(17):10079-10088. doi: 10.1093/nar/gkx654. Nucleic Acids Res. 2017. PMID: 28973450 Free PMC article.
-
The Similarities between Human Mitochondria and Bacteria in the Context of Structure, Genome, and Base Excision Repair System.Molecules. 2020 Jun 21;25(12):2857. doi: 10.3390/molecules25122857. Molecules. 2020. PMID: 32575813 Free PMC article. Review.
References
-
- Date H, et al. Early-onset ataxia with ocular motor apraxia and hypoalbuminemia is caused by mutations in a new HIT superfamily gene. Nat Genet. 2001;29:184–188. - PubMed
-
- Moreira MC, et al. The gene mutated in ataxia-ocular apraxia 1 encodes the new HIT/Zn-finger protein aprataxin. Nat Genet. 2001;29:189–193. - PubMed
-
- Ahel I, et al. The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates. Nature. 2006;443:713–716. - PubMed
-
- Kijas AW, Harris JL, Harris JM, Lavin MF. Aprataxin forms a discrete branch in the HIT (histidine triad) superfamily of proteins with both DNA/RNA binding and nucleotide hydrolase activities. J Biol Chem. 2006;281:13939–13948. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

