Conserved, developmentally regulated mechanism couples chromosomal looping and heterochromatin barrier activity at the homeobox gene A locus
- PMID: 21502535
- PMCID: PMC3088595
- DOI: 10.1073/pnas.1018279108
Conserved, developmentally regulated mechanism couples chromosomal looping and heterochromatin barrier activity at the homeobox gene A locus
Erratum in
- Proc Natl Acad Sci U S A. 2011 Nov;108(47):19096
Abstract
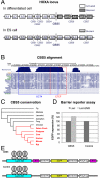
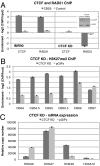
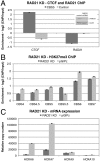
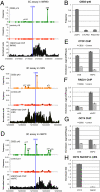
Establishment and segregation of distinct chromatin domains are essential for proper genome function. The insulator protein CCCTC-binding factor (CTCF) is involved in creating boundaries that segregate chromatin and functional domains and in organizing higher-order chromatin structures by promoting chromosomal loops across the vertebrate genome. Here, we investigate the insulation properties of CTCF at the human and mouse homeobox gene A (HOXA) loci. Although cohesin loading at the CTCF binding site is required for looping, we found that cohesin is dispensable for chromatin barrier activity at that site. Using mouse embryonic stem cells in both a pluripotent and differentiated neuronal progenitor state, we determined that embryonic stem cell pluripotency factor OCT4 antagonizes cohesin loading at the CTCF binding site. Loss of OCT4 in the committed and differentiated neuronal progenitor cells results in loading of cohesin and chromosome looping, which contributes to heterochromatin partitioning and selective gene activation across the HOXA locus. Our analysis reveals that chromatin barrier activity of CTCF is evolutionarily conserved and is responsible for the coordinated establishment of chromatin structure, higher-order architecture, and developmental expression of the HOXA locus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: Knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. - PubMed
-
- Defossez PA, et al. The human enhancer blocker CTC-binding factor interacts with the transcription factor Kaiso. J Biol Chem. 2005;280:43017–43023. - PubMed
-
- Docquier F, et al. Decreased poly(ADP-ribosyl)ation of CTCF, a transcription factor, is associated with breast cancer phenotype and cell proliferation. Clin Cancer Res. 2009;15:5762–5771. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

