Widespread transcription in an amphibian oocyte relates to its reprogramming activity on transplanted somatic nuclei
- PMID: 21504359
- PMCID: PMC3636465
- DOI: 10.1089/scd.2011.0162
Widespread transcription in an amphibian oocyte relates to its reprogramming activity on transplanted somatic nuclei
Abstract
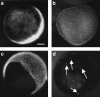
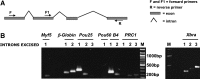
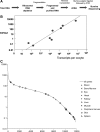
Amphibian oocytes have the special ability to directly induce the transcription of pluripotency and other genes in transplanted somatic nuclei. To this extent, oocytes induce a stem cell-like pattern of transcription in somatic cell nuclei. We ask whether the induced transcription in transplanted nuclei reflects the normal transcriptional activity of oocyte genes. We describe here the transcript content of a wide range of genes in Xenopus tropicalis oocytes. Using accurate quantitation, we find that each mature oocyte has accumulated several hundred transcripts of cell-type specific genes. This value is several orders of magnitude greater than the "leakage" level found in most somatic cells and about the same level found in somatic cells where these genes are fully expressed. Illumina sequencing confirms the high transcript content of a mature Xenopus oocyte. Most of the transcripts from these highly expressed genes in oocytes are correctly and efficiently spliced. Our results contribute a more quantitative view of certain amphibian oocyte transcripts than previously available. Our results also show that transplanted somatic nuclei conform, with respect to the genes analyzed, to the transcriptional characteristics of the recipient oocytes.
Figures

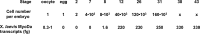
References
-
- Flemming W. Zell substanz, Kern, und Zelltheilung. F.C.W. Vogel; Leipzig: 1882.
-
- Callan HG. The nature of lampbrush chromosomes. Int Rev Cytol. 1963;15:1–34. - PubMed
-
- Callan HG. Lampbrush chromosomes. Mol Biol Biochem Biophys. 1986;36:1–252. - PubMed
-
- Callan HG. Gall JG. Berg CA. The lampbrush chromosomes of Xenopus laevis: preparation, identification, and distribution of 5S DNA sequences. Chromosoma. 1987;95:236–250. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

