PU.1-dependent regulation of UCH L1 expression in B-lymphoma cells
- PMID: 21504384
- PMCID: PMC4435811
- DOI: 10.3109/10428194.2011.562571
PU.1-dependent regulation of UCH L1 expression in B-lymphoma cells
Abstract
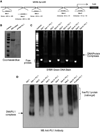
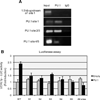
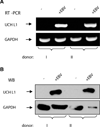
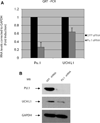
Elevated levels of ubiquitin C-terminal hydrolase L1 (UCH L1) have been detected in a variety of malignancies, and recent studies show the oncogenic capacity of overexpressed UCH L1 in vivo in animal models. Here we demonstrate that expression of endogenous UCH L1 is significantly higher in B-lymphoma cells than in transformed cells of epithelial and fibroblastic origin. The specific hematopoietic transcription factor PU.1 induces UCH L1 expression through direct activation of the uch l1 promoter. Using chromatin immunoprecipitation (ChIP) assays and direct mutagenesis we identified PU.1 binding sites on the uch l1 promoter, at least three of which are involved in this activation. We also show that the viral transcriptional co-activator EBNA2 dramatically increases PU.1-dependent up-regulation of endogenous UCH L1 expression. Finally, inhibition of PU.1 expression with specific shRNA resulted in reduction of UCH L1 mRNA and protein levels in Epstein-Barr virus (EBV)-transformed B-cells. We propose that the ubiquitin-editing enzyme UCH L1 is a multifunctional pro-oncogenic factor involved in development and progression of certain lymphoid malignancies, including EBV-associated lymphomas.
Conflict of interest statement
Figures

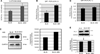

Similar articles
-
KSHV LANA and EBV LMP1 induce the expression of UCH-L1 following viral transformation.Virology. 2014 Jan 5;448:293-302. doi: 10.1016/j.virol.2013.10.018. Epub 2013 Nov 8. Virology. 2014. PMID: 24314660 Free PMC article.
-
Inhibition of UCH-L1 Deubiquitinating Activity with Two Forms of LDN-57444 Has Anti-Invasive Effects in Metastatic Carcinoma Cells.Int J Mol Sci. 2019 Jul 31;20(15):3733. doi: 10.3390/ijms20153733. Int J Mol Sci. 2019. PMID: 31370144 Free PMC article.
-
UCH-L1 is induced in germinal center B cells and identifies patients with aggressive germinal center diffuse large B-cell lymphoma.Blood. 2016 Mar 24;127(12):1564-74. doi: 10.1182/blood-2015-07-656678. Epub 2015 Dec 23. Blood. 2016. PMID: 26702068 Free PMC article.
-
The ubiquitin C-terminal hydrolase UCH-L1 regulates B-cell proliferation and integrin activation.J Cell Mol Med. 2009 Aug;13(8B):1666-1678. doi: 10.1111/j.1582-4934.2008.00501.x. J Cell Mol Med. 2009. PMID: 20187292 Free PMC article.
-
Positive reciprocal regulation of ubiquitin C-terminal hydrolase L1 and beta-catenin/TCF signaling.PLoS One. 2009 Jun 18;4(6):e5955. doi: 10.1371/journal.pone.0005955. PLoS One. 2009. PMID: 19536331 Free PMC article.
Cited by
-
Viruses and human cancers: a long road of discovery of molecular paradigms.Clin Microbiol Rev. 2014 Jul;27(3):463-81. doi: 10.1128/CMR.00124-13. Clin Microbiol Rev. 2014. PMID: 24982317 Free PMC article. Review.
-
UCH-LI acts as a novel prognostic biomarker in gastric cardiac adenocarcinoma.Int J Clin Exp Pathol. 2015 Nov 1;8(11):13957-67. eCollection 2015. Int J Clin Exp Pathol. 2015. PMID: 26823707 Free PMC article.
-
C-Terminal Farnesylation of UCH-L1 Plays a Role in Transport of Epstein-Barr Virus Primary Oncoprotein LMP1 to Exosomes.mSphere. 2018 Feb 7;3(1):e00030-18. doi: 10.1128/mSphere.00030-18. eCollection 2018 Jan-Feb. mSphere. 2018. PMID: 29435490 Free PMC article.
-
UCH-L1 in DLBCL: marker or target?Blood. 2016 Mar 24;127(12):1524-5. doi: 10.1182/blood-2016-01-689984. Blood. 2016. PMID: 27013211 Free PMC article.
-
EBF1 binds to EBNA2 and promotes the assembly of EBNA2 chromatin complexes in B cells.PLoS Pathog. 2017 Oct 2;13(10):e1006664. doi: 10.1371/journal.ppat.1006664. eCollection 2017 Oct. PLoS Pathog. 2017. PMID: 28968461 Free PMC article.
References
-
- Larsen CN, Price JS, Wilkinson KD. Substrate binding and catalysis by ubiquitin C-terminal hydrolases: identification of two active site residues. Biochemistry. 1996;35:6735–6744. - PubMed
-
- Betarbet R, Sherer TB, Greenamyre JT. Ubiquitin-proteasome system and Parkinson’s diseases. Exp Neurol. 2005;191(Suppl. 1):S17–S27. - PubMed
-
- Liu Y, Fallon L, Lashuel HA, Liu Z, Lansbury PT., Jr The UCH-L1 gene encodes two opposing enzymatic activities that affect alpha-synuclein degradation and Parkinson’s disease susceptibility. Cell. 2002;111:209–218. - PubMed
-
- Setsuie R, Wada K. The functions of UCH-L1 and its relation to neurodegenerative diseases. Neurochem Int. 2007;51:105–111. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
