Altered developmental expression of polymorphic membrane proteins in penicillin-stressed Chlamydia trachomatis
- PMID: 21504531
- PMCID: PMC3116966
- DOI: 10.1111/j.1462-5822.2011.01598.x
Altered developmental expression of polymorphic membrane proteins in penicillin-stressed Chlamydia trachomatis
Abstract
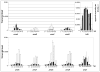
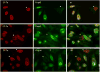
Late Chlamydia trachomatis inclusions express each member of the surface-exposed polymorphic membrane protein family (Pmp subtypes A through I) with a reproducible distribution of fully-on, fully-off and intermediate phenotypes. This observation is consistent with observed variable Pmp antibody profiles in C. trachomatis-infected patients and has led to the hypothesis that the pmp gene family forms the basis of a phase variation-like mechanism of antigenic variation. Here we investigate and compare the developmental expression of each of the nine pmp genes under conditions of optimal in vitro growth with that under conditions that promote prolonged survival of chlamydiae when exposed to penicillin-induced stress. We demonstrate that the pmp gene family includes distinct transcriptional units that are differentially expressed along development and differentially responsive to stress. In particular, our results indicate that expression of pmpA, pmpD and pmpI is uniquely unaffected by stress, suggesting that the PmpA, PmpD and PmpI proteins play a critical role in the pathogenesis of C. trachomatis.
© 2011 Blackwell Publishing Ltd.
Figures





References
-
- Al-Younes HM, Rudel T, Brinkmann V, Szczepek AJ, Meyer TF. Low iron availability modulates the course of Chlamydia pneumoniae infection. Cell Microbiol. 2001;3:427–437. - PubMed
-
- Allan I, Hatch TP, Pearce JH. Influence of cysteine deprivation on chlamydial differentiation from reproductive to infective life-cycle forms. Journal of General Microbiology. 1985;131:3171. - PubMed
-
- Allan I, Pearce JH. Differential amino acid utilization by Chlamydia psittaci (Strain guinea pig inclusion conjunctivitis) and its regulatory effect on Chlamydial growth. Journal of General Microbiology. 1983;129:1991–2000. - PubMed
-
- Azuma Y, Hirakawa H, Yamashita A, Cai Y, Rahman MA, Suzuki H, et al. Genome sequence of the cat pathogen, Chlamydophila felis. DNA Research. 2006;13:15–23. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

