Estimation of alternative splicing isoform frequencies from RNA-Seq data
- PMID: 21504602
- PMCID: PMC3107792
- DOI: 10.1186/1748-7188-6-9
Estimation of alternative splicing isoform frequencies from RNA-Seq data
Abstract
Background: Massively parallel whole transcriptome sequencing, commonly referred as RNA-Seq, is quickly becoming the technology of choice for gene expression profiling. However, due to the short read length delivered by current sequencing technologies, estimation of expression levels for alternative splicing gene isoforms remains challenging.
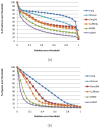
Results: In this paper we present a novel expectation-maximization algorithm for inference of isoform- and gene-specific expression levels from RNA-Seq data. Our algorithm, referred to as IsoEM, is based on disambiguating information provided by the distribution of insert sizes generated during sequencing library preparation, and takes advantage of base quality scores, strand and read pairing information when available. The open source Java implementation of IsoEM is freely available at http://dna.engr.uconn.edu/software/IsoEM/.
Conclusions: Empirical experiments on both synthetic and real RNA-Seq datasets show that IsoEM has scalable running time and outperforms existing methods of isoform and gene expression level estimation. Simulation experiments confirm previous findings that, for a fixed sequencing cost, using reads longer than 25-36 bases does not necessarily lead to better accuracy for estimating expression levels of annotated isoforms and genes.
Figures









References
-
- Griffith M, Griffith OL, Mwenifumbo J, Goya R, Morrissy AS, Morin RD, Corbett R, Tang MJ, Hou YC, Pugh TJ, Robertson G, Chittaranjan S, Ally A, Asano JK, Chan SY, Li HI, McDonald H, Teague K, Zhao Y, Zeng T, Delaney A, Hirst M, Morin GB, Jones SJM, Tai IT, Marra MA. Alternative expression analysis by RNA sequencing. Nature Methods. 2010;7(10):843–847. doi: 10.1038/nmeth.1503. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

