Jumbled genomes: missing Apicomplexan synteny
- PMID: 21504890
- PMCID: PMC3176833
- DOI: 10.1093/molbev/msr103
Jumbled genomes: missing Apicomplexan synteny
Abstract
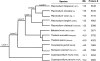
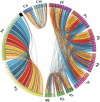
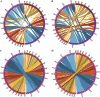
Whole-genome comparisons provide insight into genome evolution by informing on gene repertoires, gene gains/losses, and genome organization. Most of our knowledge about eukaryotic genome evolution is derived from studies of multicellular model organisms. The eukaryotic phylum Apicomplexa contains obligate intracellular protist parasites responsible for a wide range of human and veterinary diseases (e.g., malaria, toxoplasmosis, and theileriosis). We have developed an in silico protein-encoding gene based pipeline to investigate synteny across 12 apicomplexan species from six genera. Genome rearrangement between lineages is extensive. Syntenic regions (conserved gene content and order) are rare between lineages and appear to be totally absent across the phylum, with no group of three genes found on the same chromosome and in the same order within 25 kb up- and downstream of any orthologous genes. Conserved synteny between major lineages is limited to small regions in Plasmodium and Theileria/Babesia species, and within these conserved regions, there are a number of proteins putatively targeted to organelles. The observed overall lack of synteny is surprising considering the divergence times and the apparent absence of transposable elements (TEs) within any of the species examined. TEs are ubiquitous in all other groups of eukaryotes studied to date and have been shown to be involved in genomic rearrangements. It appears that there are different criteria governing genome evolution within the Apicomplexa relative to other well-studied unicellular and multicellular eukaryotes.
Figures
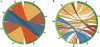
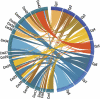
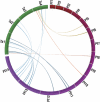
Similar articles
-
Mobilome of Apicomplexa Parasites.Genes (Basel). 2022 May 16;13(5):887. doi: 10.3390/genes13050887. Genes (Basel). 2022. PMID: 35627271 Free PMC article.
-
Divergence of the mitochondrial genome structure in the apicomplexan parasites, Babesia and Theileria.Mol Biol Evol. 2010 May;27(5):1107-16. doi: 10.1093/molbev/msp320. Epub 2009 Dec 24. Mol Biol Evol. 2010. PMID: 20034997
-
Phylogeny and evolution of apicoplasts and apicomplexan parasites.Parasitol Int. 2015 Jun;64(3):254-9. doi: 10.1016/j.parint.2014.10.005. Epub 2014 Oct 14. Parasitol Int. 2015. PMID: 25451217 Review.
-
Diversity of mitochondrial genome structure in the phylum Apicomplexa.Mol Biochem Parasitol. 2013 Mar;188(1):26-33. doi: 10.1016/j.molbiopara.2013.02.006. Epub 2013 Mar 4. Mol Biochem Parasitol. 2013. PMID: 23466751 Review.
-
Genomic hotspots of chromosome rearrangements explain conserved synteny despite high rates of chromosome evolution in a holocentric lineage.Mol Ecol. 2024 Dec;33(24):e17086. doi: 10.1111/mec.17086. Epub 2023 Jul 24. Mol Ecol. 2024. PMID: 37486041 Free PMC article.
Cited by
-
Comparative genomics of the apicomplexan parasites Toxoplasma gondii and Neospora caninum: Coccidia differing in host range and transmission strategy.PLoS Pathog. 2012;8(3):e1002567. doi: 10.1371/journal.ppat.1002567. Epub 2012 Mar 22. PLoS Pathog. 2012. PMID: 22457617 Free PMC article.
-
Mobilome of Apicomplexa Parasites.Genes (Basel). 2022 May 16;13(5):887. doi: 10.3390/genes13050887. Genes (Basel). 2022. PMID: 35627271 Free PMC article.
-
An evolutionary perspective on the kinome of malaria parasites.Philos Trans R Soc Lond B Biol Sci. 2012 Sep 19;367(1602):2607-18. doi: 10.1098/rstb.2012.0014. Philos Trans R Soc Lond B Biol Sci. 2012. PMID: 22889911 Free PMC article. Review.
-
The HU protein is important for apicoplast genome maintenance and inheritance in Toxoplasma gondii.Eukaryot Cell. 2012 Jul;11(7):905-15. doi: 10.1128/EC.00029-12. Epub 2012 May 18. Eukaryot Cell. 2012. PMID: 22611021 Free PMC article.
-
Comparative genomic analysis and phylogenetic position of Theileria equi.BMC Genomics. 2012 Nov 9;13:603. doi: 10.1186/1471-2164-13-603. BMC Genomics. 2012. PMID: 23137308 Free PMC article.
References
-
- Abrahamsen MS, Templeton TJ, Enomoto S, et al. (20 co-authors) Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science. 2004;304:441–445. - PubMed
-
- al-Khedery B, Barnwell JW, Galinski MR. Antigenic variation in malaria: a 3' genomic alteration associated with the expression of a P. knowlesi variant antigen. Mol Cell. 1999;3:131–141. - PubMed
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

