Global hypomethylation identifies Loci targeted for hypermethylation in head and neck cancer
- PMID: 21505061
- PMCID: PMC3107873
- DOI: 10.1158/1078-0432.CCR-11-0044
Global hypomethylation identifies Loci targeted for hypermethylation in head and neck cancer
Abstract
Purpose: The human epigenome is profoundly altered in cancers, with a characteristic loss of methylation in repetitive regions and concomitant accumulation of gene promoter methylation. The degree to which these processes are coordinated is unclear so we investigated both in head and neck squamous cell carcinomas.
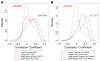
Experimental design: Global methylation was measured using the luminometric methylation assay (LUMA) and pyrosequencing of LINE-1Hs and AluYb8 repetitive elements in a series of 138 tumors. We also measured methylation of more than 27,000 CpG loci with the Illumina HumanMethylation27 Microarray (n = 91).
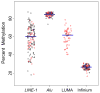
Results: LINE-1 methylation was significantly associated with LUMA and Infinium loci methylation (Spearman's ρ = 0.52/ρ = 0.56, both P < 0.001) but not that of AluYb8. Methylation of LINE-1, AluYb8, and Infinium loci differed by tumor site (each Kruskal-Wallis, P < 0.05). Also, LINE-1 and LUMA methylation were associated with HPV16 E6 serology (each Mann-Whitney, P < 0.05). Comparing LINE-1 methylation to gene-associated methylation, we identified a distinct subset of CpG loci with significant hypermethylation associated with LINE-1 hypomethylation. An investigation of sequence features for these CpG loci revealed that they were significantly less likely to reside in repetitive elements (Gene Set Enrichment Analysis, P < 0.02), enriched in CpG islands (P < 0.001) and were proximal to transcription factor-binding sites (P < 0.05). We validated the top CpG loci that had significant hypermethylation associated with LINE-1 hypomethylation (at EVI2A, IFRD1, KLHL6, and PTPRCAP) by pyrosequencing independent tumors.
Conclusions: These data indicate that global hypomethylation and gene-specific methylation processes are associated in a sequence-dependent manner, and that clinical characteristics and exposures leading to HNSCC may be influencing these processes.
©2011 AACR.
Conflict of interest statement
The authors declare no potential conflicts of interest.
Figures


References
-
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–28. - PubMed
-
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–49. - PubMed
-
- Hasegawa M, Nelson HH, Peters E, Ringstrom E, Posner M, Kelsey KT. Patterns of gene promoter methylation in squamous cell cancer of the head and neck. Oncogene. 2002;21:4231–6. - PubMed
-
- Dikshit RP, Gillio-Tos A, Brennan P, et al. Hypermethylation, risk factors, clinical characteristics, and survival in 235 patients with laryngeal and hypopharyngeal cancers. Cancer. 2007;110:1745–51. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

