Common and overlapping oncogenic pathways contribute to the evolution of acute myeloid leukemias
- PMID: 21505102
- PMCID: PMC3119437
- DOI: 10.1158/0008-5472.CAN-11-0176
Common and overlapping oncogenic pathways contribute to the evolution of acute myeloid leukemias
Abstract
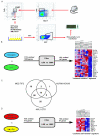
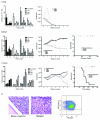
Fusion oncogenes in acute myeloid leukemia (AML) promote self-renewal from committed progenitors, thereby linking transformation and self-renewal pathways. Like most cancers, AML is a genetically and biologically heterogeneous disease, but it is unclear whether transformation results from common or overlapping genetic programs acting downstream of multiple mutations or by the engagement of unique genetic programs acting cooperatively downstream of individual mutations. This distinction is important, because the involvement of common programs would imply the existence of common molecular targets to treat AML, no matter which oncogenes are involved. Here we show that the ability to promote self-renewal is a generalized property of leukemia-associated oncogenes. Disparate oncogenes initiated overlapping transformation and self-renewal gene expression programs, the common elements of which were defined in established leukemic stem cells from an animal model as well as from a large cohort of patients with differing AML subtypes, where they strongly predicted pathobiological character. Notably, individual genes commonly activated in these programs could partially phenocopy the self-renewal function of leukemia-associated oncogenes in committed murine progenitors. Furthermore, they could generate AML following expression in murine bone marrow. In summary, our findings reveal the operation of common programs of self-renewal and transformation downstream of leukemia-associated oncogenes, suggesting that mechanistically common therapeutic approaches to AML are likely to be possible, regardless of the identity of the driver oncogene involved.
Figures



References
-
- Estey E, Dohner H. Acute myeloid leukaemia. Lancet. 2006;368:1894–1907. - PubMed
-
- Bullinger L, Dohner K, Bair E, et al. Use of gene-expression profiling to identify prognostic subclasses in adult acute myeloid leukemia. N Engl J Med. 2004;350:1605–1616. - PubMed
-
- Valk PJ, Verhaak RG, Beijen MA, et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med. 2004;350:1617–1628. - PubMed
-
- Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

