Use of whole-genome sequencing to diagnose a cryptic fusion oncogene
- PMID: 21505136
- PMCID: PMC3156695
- DOI: 10.1001/jama.2011.497
Use of whole-genome sequencing to diagnose a cryptic fusion oncogene
Abstract
Context: Whole-genome sequencing is becoming increasingly available for research purposes, but it has not yet been routinely used for clinical diagnosis.
Objective: To determine whether whole-genome sequencing can identify cryptic, actionable mutations in a clinically relevant time frame.
Design, setting, and patient: We were referred a difficult diagnostic case of acute promyelocytic leukemia with no pathogenic X-RARA fusion identified by routine metaphase cytogenetics or interphase fluorescence in situ hybridization (FISH). The case patient was enrolled in an institutional review board-approved protocol, with consent specifically tailored to the implications of whole-genome sequencing. The protocol uses a "movable firewall" that maintains patient anonymity within the entire research team but allows the research team to communicate medically relevant information to the treating physician.
Main outcome measures: Clinical relevance of whole-genome sequencing and time to communicate validated results to the treating physician.
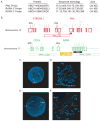
Results: Massively parallel paired-end sequencing allowed identification of a cytogenetically cryptic event: a 77-kilobase segment from chromosome 15 was inserted en bloc into the second intron of the RARA gene on chromosome 17, resulting in a classic bcr3 PML-RARA fusion gene. Reverse transcription polymerase chain reaction sequencing subsequently validated the expression of the fusion transcript. Novel FISH probes identified 2 additional cases of t(15;17)-negative acute promyelocytic leukemia that had cytogenetically invisible insertions. Whole-genome sequencing and validation were completed in 7 weeks and changed the treatment plan for the patient.
Conclusion: Whole-genome sequencing can identify cytogenetically invisible oncogenes in a clinically relevant time frame.
Figures


Comment in
-
Whole-genome sequencing: a step closer to personalized medicine.JAMA. 2011 Apr 20;305(15):1596-7. doi: 10.1001/jama.2011.484. JAMA. 2011. PMID: 21505140 No abstract available.
-
Whole-genome sequencing and acute promyelocytic leukemia.JAMA. 2011 Aug 10;306(6):610; author reply 610-1. doi: 10.1001/jama.2011.1108. JAMA. 2011. PMID: 21828321 No abstract available.
References
-
- Jurcic JG, Soignet SL, Maslak AP. Diagnosis and treatment of acute promyelocytic leukemia. Curr Oncol Rep. 2007 Sep;9(5):337–344. - PubMed
-
- Grimwade D, Biondi A, Mozziconacci MJ, et al. Characterization of acute promyelocytic leukemia cases lacking the classic t(15;17): results of the European Working Party. Groupe Francais de Cytogenetique Hematologique, Groupe de Francais d’Hematologie Cellulaire, UK Cancer Cytogenetics Group and BIOMED 1 European Community-Concerted Action “Molecular Cytogenetic Diagnosis in Haematological Malignancies”. Blood. 2000 Aug 15;96(4):1297–1308. - PubMed
-
- Abe S, Ishikawa I, Harigae H, Sugawara T. A new complex translocation t(5;17;15)(q11;q21;q22) in acute promyelocytic leukemia. Cancer Genet Cytogenet. 2008 Jul;184(1):44–47. - PubMed
-
- Hiorns LR, Swansbury GJ, Catovsky D. An eight-way variant t(15;17) in acute promyelocytic leukemia elucidated using fluorescence in situ hybridization. Cancer Genet Cytogenet. 1995 Sep;83(2):136–139. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

