Extended high viremics: a substantial fraction of individuals maintain high plasma viral RNA levels after acute HIV-1 subtype C infection
- PMID: 21505307
- PMCID: PMC3544358
- DOI: 10.1097/QAD.0b013e3283471eb2
Extended high viremics: a substantial fraction of individuals maintain high plasma viral RNA levels after acute HIV-1 subtype C infection
Abstract
Objective: The present study addressed two questions: what fraction of individuals maintain a sustained high HIV-1 RNA load after the acute HIV-1C infection peak and how long is a high HIV-1 RNA load maintained after acute HIV-1C infection in this subpopulation?
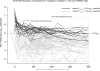
Design/methods: Plasma HIV-1 RNA dynamics were studied in 77 participants with primary HIV-1C infection from African cohorts in Gaborone, Botswana, and Durban, South Africa. HIV-infected individuals who maintained mean viral load of at least 100,000 (5.0 log(10)) copies/ml after 100 days postseroconversion (p/s) were termed extended high viremics. Individuals were followed longitudinally for a median [interquartile range (IQR)] of 573 (226-986) days p/s.
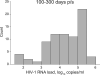
Results: The proportion of extended high viremics was 34% [95% confidence interval (CI) 23-44%] during the period 100-300 days p/s and 19% (95% CI 9-29%) over the period of 200-400 days p/s. The median (IQR) duration of HIV-1 RNA load at least 100,000 copies/ml among extended high viremics was 271 (188-340) days p/s. For the subset with average viral load at least 100,000 copies/ml during 200-400 days p/s, the median (IQR) duration was 318 (282-459) days. The extended high viremics had a significantly shorter time to CD4 cell decline to 350 cells/μl (median: 88 vs. 691 days p/s for those not designated as extended high viremics; P < 0.0001, Gehan-Wilcoxon test).
Conclusion: A high proportion of extended high viremics - individuals maintaining high plasma HIV-1 RNA load after acute infection - have been identified during primary HIV-1 subtype C infection. These extended high viremics likely contribute disproportionately to HIV-1 incidence.
Figures



Comment in
-
High clade C HIV-1 viremia: how did we get here and where are we going?AIDS. 2011 Jul 31;25(12):1543-5. doi: 10.1097/QAD.0b013e328348a4d2. AIDS. 2011. PMID: 21747235 No abstract available.
References
-
- Kaufmann GR, Cunningham P, Kelleher AD, Zaunders J, Carr A, Vizzard J, et al. Patterns of viral dynamics during primary human immunodeficiency virus type 1 infection. The Sydney Primary HIV Infection Study Group. J Infect Dis. 1998;178:1812–1815. - PubMed
-
- Kaufmann GR, Duncombe C, Zaunders J, Cunningham P, Cooper D. Primary HIV-1 infection: a review of clinical manifestations, immunologic and virologic changes. AIDS Patient Care STDS. 1998;12:759–767. - PubMed
-
- Fiebig EW, Wright DJ, Rawal BD, Garrett PE, Schumacher RT, Peddada L, et al. Dynamics of HIV viremia and antibody seroconversion in plasma donors: implications for diagnosis and staging of primary HIV infection. AIDS. 2003;17:1871–1879. - PubMed
-
- Schacker TW, Hughes JP, Shea T, Coombs RW, Corey L. Biological and virologic characteristics of primary HIV infection. Ann Intern Med. 1998;128:613–620. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

