PheMaDB: a solution for storage, retrieval, and analysis of high throughput phenotype data
- PMID: 21507258
- PMCID: PMC3097161
- DOI: 10.1186/1471-2105-12-109
PheMaDB: a solution for storage, retrieval, and analysis of high throughput phenotype data
Abstract
Background: OmniLog™ phenotype microarrays (PMs) have the capability to measure and compare the growth responses of biological samples upon exposure to hundreds of growth conditions such as different metabolites and antibiotics over a time course of hours to days. In order to manage the large amount of data produced from the OmniLog™ instrument, PheMaDB (Phenotype Microarray DataBase), a web-based relational database, was designed. PheMaDB enables efficient storage, retrieval and rapid analysis of the OmniLog™ PM data.
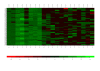
Description: PheMaDB allows the user to quickly identify records of interest for data analysis by filtering with a hierarchical ordering of Project, Strain, Phenotype, Replicate, and Temperature. PheMaDB then provides various statistical analysis options to identify specific growth pattern characteristics of the experimental strains, such as: outlier analysis, negative controls analysis (signal/background calibration), bar plots, pearson's correlation matrix, growth curve profile search, k-means clustering, and a heat map plot. This web-based database management system allows for both easy data sharing among multiple users and robust tools to phenotype organisms of interest.
Conclusions: PheMaDB is an open source system standardized for OmniLog™ PM data. PheMaDB could facilitate the banking and sharing of phenotype data. The source code is available for download at http://phemadb.sourceforge.net.
Figures


References
-
- R Development Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. 2009. ISBN 3-900051-07-0.
-
- Sozhamannan S, McKinstry M, Lentz SM, Jalasvuori M, McAfee F, Smith A, Dabbs J, Ackermann HW, Bamford JK, Mateczun A, Read TD. Molecular characterization of a variant of Bacillus anthracis-specific phage AP50 with improved bacteriolytic activity. Appl Environ Microbiol. 2008;74(21):6792–6. doi: 10.1128/AEM.01124-08. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

