A disease-associated polymorphism alters splicing of the human CD45 phosphatase gene by disrupting combinatorial repression by heterogeneous nuclear ribonucleoproteins (hnRNPs)
- PMID: 21507955
- PMCID: PMC3103377
- DOI: 10.1074/jbc.M111.218727
A disease-associated polymorphism alters splicing of the human CD45 phosphatase gene by disrupting combinatorial repression by heterogeneous nuclear ribonucleoproteins (hnRNPs)
Abstract
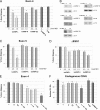
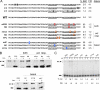
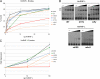
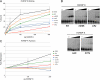
Alternative splicing is typically controlled by complexes of regulatory proteins that bind to sequences within or flanking variable exons. The identification of regulatory sequence motifs and the characterization of sequence motifs bound by splicing regulatory proteins have been essential to predicting splicing regulation. The activation-responsive sequence (ARS) motif has previously been identified in several exons that undergo changes in splicing upon T cell activation. hnRNP L binds to this ARS motif and regulates ARS-containing exons; however, hnRNP L does not function alone. Interestingly, the proteins that bind together with hnRNP L differ for different exons that contain the ARS core motif. Here we undertake a systematic mutational analysis of the best characterized context of the ARS motif, namely the ESS1 sequence from CD45 exon 4, to understand the determinants of binding specificity among the components of the ESS1 regulatory complex and the relationship between protein binding and function. We demonstrate that different mutations within the ARS motif affect specific aspects of regulatory function and disrupt the binding of distinct proteins. Most notably, we demonstrate that the C77G polymorphism, which correlates with autoimmune disease susceptibility in humans, disrupts exon silencing by preventing the redundant activity of hnRNPs K and E2 to compensate for the weakened function of hnRNP L. Therefore, these studies provide an important example of the functional relevance of combinatorial function in splicing regulation and suggest that additional polymorphisms may similarly disrupt function of the ESS1 silencer.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

