Blocking eIF4E-eIF4G interaction as a strategy to impair coronavirus replication
- PMID: 21507972
- PMCID: PMC3126520
- DOI: 10.1128/JVI.00078-11
Blocking eIF4E-eIF4G interaction as a strategy to impair coronavirus replication
Abstract
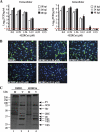
Coronaviruses are a family of enveloped single-stranded positive-sense RNA viruses causing respiratory, enteric, and neurologic diseases in mammals and fowl. Human coronaviruses are recognized to cause up to a third of common colds and are suspected to be involved in enteric and neurologic diseases. Coronavirus replication involves the generation of nested subgenomic mRNAs (sgmRNAs) with a common capped 5' leader sequence. The translation of most of the sgmRNAs is thought to be cap dependent and displays a requirement for eukaryotic initiation factor 4F (eIF4F), a heterotrimeric complex needed for the recruitment of 40S ribosomes. We recently reported on an ultrahigh-throughput screen to discover compounds that inhibit eIF4F activity by blocking the interaction of two of its subunits (R. Cencic et al., Proc. Natl. Acad. Sci. U. S. A. 108:1046-1051, 2011). Herein we describe a molecule from this screen that prevents the interaction between eIF4E (the cap-binding protein) and eIF4G (a large scaffolding protein), inhibiting cap-dependent translation. This inhibitor significantly decreased human coronavirus 229E (HCoV-229E) replication, reducing the percentage of infected cells and intra- and extracellular infectious virus titers. Our results support the strategy of targeting the eIF4F complex to block coronavirus infection.
Figures

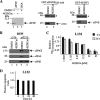
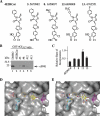
References
-
- Bordeleau M.-E., et al. 2006. Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nat. Chem. Biol. 2:213–220 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

