A yeast model for polyalanine-expansion aggregation and toxicity
- PMID: 21508314
- PMCID: PMC3113764
- DOI: 10.1091/mbc.E11-01-0037
A yeast model for polyalanine-expansion aggregation and toxicity
Abstract
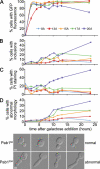
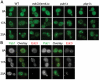
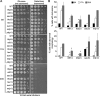
Nine human disorders result from the toxic accumulation and aggregation of proteins with expansions in their endogenous polyalanine (polyA) tracts. Given the prevalence of polyA tracts in eukaryotic proteomes, we wanted to understand the generality of polyA-expansion cytotoxicity by using yeast as a model organism. In our initial case, we expanded the polyA tract within the native yeast poly(Adenine)-binding protein Pab1 from 8A to 13A, 15A, 17A, and 20A. These expansions resulted in increasing formation of Pab1 inclusions, insolubility, and cytotoxicity that correlated with the length of the polyA expansion. Pab1 binds mRNA as part of its normal function, and disrupting RNA binding or altering cytoplasmic mRNA levels suppressed the cytotoxicity of 17A-expanded Pab1, indicating a requisite role for mRNA in Pab1 polyA-expansion toxicity. Surprisingly, neither manipulation suppressed the cytotoxicity of 20A-expanded Pab1. Thus longer expansions may have a different mechanism for toxicity. We think that this difference underscores the potential need to examine the cytotoxic mechanisms of both long and short expansions in models of expansion disorders.
Figures

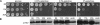






References
-
- Abu-Baker A, Messaed C, Laganiere J, Gaspar C, Brais B, Rouleau GA. Involvement of the ubiquitin-proteasome pathway and molecular chaperones in oculopharyngeal muscular dystrophy. Hum Mol Genet. 2003;12:2609–2623. - PubMed
-
- Albrecht A, Mundlos S. The other trinucleotide repeat: polyalanine expansion disorders. Curr Opin Genet Dev. 2005;15:285–293. - PubMed
-
- Albrecht AN, Kornak U, Boddrich A, Suring K, Robinson PN, Stiege AC, Lurz R, Stricker S, Wanker EE, Mundlos S. A molecular pathogenesis for transcription factor associated poly-alanine tract expansions. Hum Mol Genet. 2004;13:2351–2359. - PubMed
-
- Amiel J, Trochet D, Clement-Ziza M, Munnich A, Lyonnet S. Polyalanine expansions in human. Hum Mol Genet. 2004;13 Spec No 2:R235–R243. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

