Centromeres: unique chromatin structures that drive chromosome segregation
- PMID: 21508988
- PMCID: PMC3288958
- DOI: 10.1038/nrm3107
Centromeres: unique chromatin structures that drive chromosome segregation
Abstract
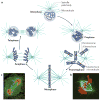
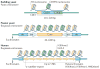
Fidelity during chromosome segregation is essential to prevent aneuploidy. The proteins and chromatin at the centromere form a unique site for kinetochore attachment and allow the cell to sense and correct errors during chromosome segregation. Centromeric chromatin is characterized by distinct chromatin organization, epigenetics, centromere-associated proteins and histone variants. These include the histone H3 variant centromeric protein A (CENPA), the composition and deposition of which have been widely investigated. Studies have examined the structural and biophysical properties of the centromere and have suggested that the centromere is not simply a 'landing pad' for kinetochore formation, but has an essential role in mitosis by assembling and directing the organization of the kinetochore.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



References
-
- Cheeseman IM, Desai A. Molecular architecture of the kinetochore–microtubule interface. Nature Rev Mol Cell Biol. 2008;9:33–46. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

