X-ray absorption spectroscopy structural investigation of early intermediates in the mechanism of DNA repair by human ABH2
- PMID: 21510633
- PMCID: PMC3124014
- DOI: 10.1021/bi101668x
X-ray absorption spectroscopy structural investigation of early intermediates in the mechanism of DNA repair by human ABH2
Abstract
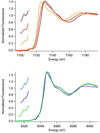
Human ABH2 repairs DNA lesions by using an Fe(II)- and αKG-dependent oxidative demethylation mechanism. The structure of the active site features the facial triad of protein ligands consisting of the side chains of two histidine residues and one aspartate residue that is common to many non-heme Fe(II) oxygenases. X-ray absorption spectroscopy (XAS) of metallated (Fe and Ni) samples of ABH2 was used to investigate the mechanism of ABH2 and its inhibition by Ni(II) ions. The data are consistent with a sequential mechanism that features a five-coordinate metal center in the presence and absence of the α-ketoglutarate cofactor. This aspect is not altered in the Ni(II)-substituted enzyme, and both metals are shown to bind the cofactor. When the substrate is bound to the native Fe(II) complex with α-ketoglutarate bound, a five-coordinate Fe(II) center is retained that features an open coordination position for O(2) binding. However, in the case of the Ni(II)-substituted enzyme, the complex that forms in the presence of the cofactor and substrate is six-coordinate and, therefore, features no open coordination site for oxygen activation at the metal.
Figures






References
-
- Martienssen RA, Colot V. DNA methylation and epigenetic inheritance in plants and filamentous fungi. Science. 2001;293:1070–1074. - PubMed
-
- Bird A. DNA methylation patterns and epigenetic memory. Gene Dev. 2002;16:6–21. - PubMed
-
- Tariq M, Paszkowski J. DNA and histone methylation in plants. Trends Genet. 2004;20:244–251. - PubMed
-
- Bender J. DNA methylation and epigenetics. Annu. Rev. Plant Biol. 2004;55:41–68. - PubMed
-
- Chan SWL, Henderson IR, Jacobsen SE. Gardening the genome: DNA methylation in arabidopsis thaliana. Nat. Rev. Genet. 2005;6:351–360. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

