Metalloregulatory proteins: metal selectivity and allosteric switching
- PMID: 21511390
- PMCID: PMC3097251
- DOI: 10.1016/j.bpc.2011.03.010
Metalloregulatory proteins: metal selectivity and allosteric switching
Abstract
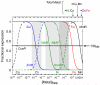
Prokaryotic organisms have evolved the capacity to quickly adapt to a changing and challenging microenvironment in which the availability of both biologically required and non-essential transition metal ions can vary dramatically. In all bacteria, a panel of metalloregulatory proteins controls the expression of genes encoding membrane transporters and metal trafficking proteins that collectively manage metal homeostasis and resistance. These "metal sensors" are specialized allosteric proteins, in which the direct binding of a specific or small number of "cognate" metal ion(s) drives a conformational change in the regulator that allosterically activates or inhibits operator DNA binding, or alternatively, distorts the promoter structure thereby converting a poor promoter to a strong one. In this review, we discuss our current understanding of the features that control metal specificity of the allosteric response in these systems, and the role that structure, thermodynamics and conformational dynamics play in mediating allosteric activation or inhibition of DNA binding.
Copyright © 2011 Elsevier B.V. All rights reserved.
Figures

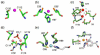

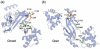


References
-
- Berg JM, Godwin HA. Lessons from zinc-binding peptides. Annu Rev Biophys Biomol Struct. 1997;26:357–371. - PubMed
-
- Seneque O, Bonnet E, Joumas FL, Latour JM. Cooperative Metal Binding and Helical Folding in Model Peptides of Treble-Clef Zinc Fingers. Chemistry. 2009;15:4798–4810. - PubMed
-
- Auld DS. Zinc coordination sphere in biochemical zinc sites. Biometals. 2001;14:271–313. - PubMed
-
- Ermler U, Grabarse W, Shima S, Goubeaud M, Thauer RK. Active sites of transition-metal enzymes with a focus on nickel. Curr Opin Struct Biol. 1998;8:749–758. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

