The proteogenomic mapping tool
- PMID: 21513508
- PMCID: PMC3107813
- DOI: 10.1186/1471-2105-12-115
The proteogenomic mapping tool
Abstract
Background: High-throughput mass spectrometry (MS) proteomics data is increasingly being used to complement traditional structural genome annotation methods. To keep pace with the high speed of experimental data generation and to aid in structural genome annotation, experimentally observed peptides need to be mapped back to their source genome location quickly and exactly. Previously, the tools to do this have been limited to custom scripts designed by individual research groups to analyze their own data, are generally not widely available, and do not scale well with large eukaryotic genomes.
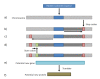
Results: The Proteogenomic Mapping Tool includes a Java implementation of the Aho-Corasick string searching algorithm which takes as input standardized file types and rapidly searches experimentally observed peptides against a given genome translated in all 6 reading frames for exact matches. The Java implementation allows the application to scale well with larger eukaryotic genomes while providing cross-platform functionality.
Conclusions: The Proteogenomic Mapping Tool provides a standalone application for mapping peptides back to their source genome on a number of operating system platforms with standard desktop computer hardware and executes very rapidly for a variety of datasets. Allowing the selection of different genetic codes for different organisms allows researchers to easily customize the tool to their own research interests and is recommended for anyone working to structurally annotate genomes using MS derived proteomics data.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

