c-Raf, but not B-Raf, is essential for development of K-Ras oncogene-driven non-small cell lung carcinoma
- PMID: 21514245
- PMCID: PMC4854330
- DOI: 10.1016/j.ccr.2011.04.002
c-Raf, but not B-Raf, is essential for development of K-Ras oncogene-driven non-small cell lung carcinoma
Abstract
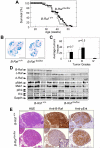
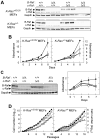
We have investigated the role of individual members of the Raf/Mek/Erk cascade in the onset of K-Ras oncogene-driven non-small cell lung carcinoma (NSCLC). Ablation of Erk1 or Erk2 in K-Ras oncogene-expressing lung cells had no significant effect due to compensatory activities. Yet, elimination of both Erk kinases completely blocked tumor development. Similar results were obtained with Mek kinases. Ablation of B-Raf had no significant effect on tumor development. However, c-Raf expression was absolutely essential for the onset of NSCLC. Interestingly, concomitant elimination of c-Raf and B-Raf in adult mice had no deleterious consequences for normal homeostasis. These results indicate that c-Raf plays a unique role in mediating K-Ras signaling and makes it a suitable target for therapeutic intervention.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

