NEMO/NLK phosphorylates PERIOD to initiate a time-delay phosphorylation circuit that sets circadian clock speed
- PMID: 21514639
- PMCID: PMC3092788
- DOI: 10.1016/j.cell.2011.04.002
NEMO/NLK phosphorylates PERIOD to initiate a time-delay phosphorylation circuit that sets circadian clock speed
Erratum in
- Cell. 2011 May13;145(4):635
Abstract
The speed of circadian clocks in animals is tightly linked to complex phosphorylation programs that drive daily cycles in the levels of PERIOD (PER) proteins. Using Drosophila, we identify a time-delay circuit based on hierarchical phosphorylation that controls the daily downswing in PER abundance. Phosphorylation by the NEMO/NLK kinase at the "per-short" domain on PER stimulates phosphorylation by DOUBLETIME (DBT/CK1δ/ɛ) at several nearby sites. This multisite phosphorylation operates in a spatially oriented and graded manner to delay progressive phosphorylation by DBT at other more distal sites on PER, including those required for recognition by the F box protein SLIMB/β-TrCP and proteasomal degradation. Highly phosphorylated PER has a more open structure, suggesting that progressive increases in global phosphorylation contribute to the timing mechanism by slowly increasing PER susceptibility to degradation. Our findings identify NEMO as a clock kinase and demonstrate that long-range interactions between functionally distinct phospho-clusters collaborate to set clock speed.
Copyright © 2011 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




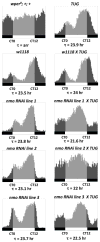
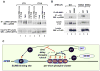
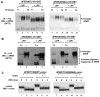
References
-
- Bae K, Edery I. Regulating a circadian clock's period, phase and amplitude by phosphorylation: insights from Drosophila. J Biochem. 2006;140:609–617. - PubMed
-
- Baylies MK, Bargiello TA, Jackson FR, Young MW. Changes in abundance or structure of the per gene product can alter periodicity of the Drosophila clock. Nature. 1987;326:390–392. - PubMed
-
- Baylies MK, Vosshall LB, Sehgal A, Young MW. New short period mutations of the Drosophila clock gene per. Neuron. 1992;9:575–581. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

