Transcriptional profiling of azole-resistant Candida parapsilosis strains
- PMID: 21518843
- PMCID: PMC3122401
- DOI: 10.1128/AAC.01127-10
Transcriptional profiling of azole-resistant Candida parapsilosis strains
Abstract
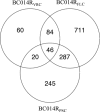
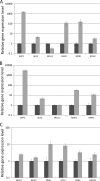
Herein we describe the changes in the gene expression profile of Candida parapsilosis associated with the acquisition of experimentally induced resistance to azole antifungal drugs. Three resistant strains of C. parapsilosis were obtained following prolonged in vitro exposure of a susceptible clinical isolate to constant concentrations of fluconazole, voriconazole, or posaconazole. We found that after incubation with fluconazole or voriconazole, strains became resistant to both azoles but not to posaconazole, although susceptibility to this azole decreased, whereas the strain incubated with posaconazole displayed resistance to the three azoles. The resistant strains obtained after exposure to fluconazole and to voriconazole have increased expression of the transcription factor MRR1, the major facilitator transporter MDR1, and several reductases and oxidoreductases. Interestingly, and similarly to what has been described in C. albicans, upregulation of MRR1 and MDR1 is correlated with point mutations in MRR1 in the resistant strains. The resistant strain obtained after exposure to posaconazole shows upregulation of two transcription factors (UPC2 and NDT80) and increased expression of 13 genes involved in ergosterol biosynthesis. This is the first study addressing global molecular mechanisms underlying azole resistance in C. parapsilosis; the results suggest that similarly to C. albicans, tolerance to azoles involves the activation of efflux pumps and/or increased ergosterol synthesis.
Figures


References
-
- Akins R. A. 2005. An update on antifungal targets and mechanisms of resistance in Candida albicans. Med. Mycol. 43:285–318 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

