Hepatocyte-targeted expression by integrase-defective lentiviral vectors induces antigen-specific tolerance in mice with low genotoxic risk
- PMID: 21520180
- PMCID: PMC3112259
- DOI: 10.1002/hep.24230
Hepatocyte-targeted expression by integrase-defective lentiviral vectors induces antigen-specific tolerance in mice with low genotoxic risk
Abstract
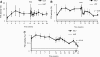
Lentiviral vectors are attractive tools for liver-directed gene therapy because of their capacity for stable gene expression and the lack of preexisting immunity in most human subjects. However, the use of integrating vectors may raise some concerns about the potential risk of insertional mutagenesis. Here we investigated liver gene transfer by integrase-defective lentiviral vectors (IDLVs) containing an inactivating mutation in the integrase (D64V). Hepatocyte-targeted expression using IDLVs resulted in the sustained and robust induction of immune tolerance to both intracellular and secreted proteins, despite the reduced transgene expression levels in comparison with their integrase-competent vector counterparts. IDLV-mediated and hepatocyte-targeted coagulation factor IX (FIX) expression prevented the induction of neutralizing antibodies to FIX even after antigen rechallenge in hemophilia B mice and accounted for relatively prolonged therapeutic FIX expression levels. Upon the delivery of intracellular model antigens, hepatocyte-targeted IDLVs induced transgene-specific regulatory T cells that contributed to the observed immune tolerance. Deep sequencing of IDLV-transduced livers showed only rare genomic integrations that had no preference for gene coding regions and occurred mostly by a mechanism inconsistent with residual integrase activity.
Conclusion: IDLVs provide an attractive platform for the tolerogenic expression of intracellular or secreted proteins in the liver with a substantially reduced risk of insertional mutagenesis.
Copyright © 2011 American Association for the Study of Liver Diseases.
Figures



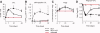


References
-
- Nguyen TH, Ferry N. Liver gene therapy: advances and hurdles. Gene Ther. 2004;11(Suppl 1):S76–S84. - PubMed
-
- Manno CS, Pierce GF, Arruda VR, Glader B, Ragni M, Rasko JJ, et al. Successful transduction of liver in hemophilia by AAV-factor IX and limitations imposed by the host immune response. Nat Med. 2006;12:342–347. - PubMed
-
- Pierce GF, Lillicrap D, Pipe SW, Vandendriessche T. Gene therapy, bioengineered clotting factors and novel technologies for hemophilia treatment. J Thromb Haemost. 2007;5:901–906. - PubMed
-
- Wasserfall CH, Herzog RW. Gene therapy approaches to induce tolerance in autoimmunity by reshaping the immune system. Curr Opin Investig Drugs. 2009;10:1143–1150. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
