Active-site-directed chemical tools for profiling mitochondrial Lon protease
- PMID: 21520912
- PMCID: PMC3158820
- DOI: 10.1021/cb100408w
Active-site-directed chemical tools for profiling mitochondrial Lon protease
Abstract
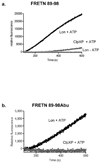
Lon and ClpXP are the only soluble ATP-dependent proteases within the mammalian mitochondria matrix, which function in protein quality control by selectively degrading misfolded, misassembled, or damaged proteins. Chemical tools to study these proteases in biological samples have not been identified, thereby hindering a clear understanding of their respective functions in normal and disease states. In this study, we applied a proteolytic site-directed approach to identify a peptide reporter substrate and a peptide inhibitor that are selective for Lon but not ClpXP. These chemical tools permit quantitative measurements that distinguish Lon-mediated proteolysis from that of ClpXP in biochemical assays with purified proteases, as well as in intact mitochondria and mitochondrial lysates. This chemical biology approach provides needed tools to further our understanding of mitochondrial ATP-dependent proteolysis and contributes to the future development of diagnostic and pharmacological agents for treating diseases associated with defects in mitochondrial protein quality.
Figures





Similar articles
-
A Selective Fluorogenic Peptide Substrate for the Human Mitochondrial ATP-Dependent Protease Complex ClpXP.Chembiochem. 2020 Jul 16;21(14):2037-2048. doi: 10.1002/cbic.202000030. Epub 2020 Apr 2. Chembiochem. 2020. PMID: 32180333
-
LON is the master protease that protects against protein aggregation in human mitochondria through direct degradation of misfolded proteins.Sci Rep. 2015 Dec 2;5:17397. doi: 10.1038/srep17397. Sci Rep. 2015. PMID: 26627475 Free PMC article.
-
Matrix proteases in mitochondrial DNA function.Biochim Biophys Acta. 2012 Sep-Oct;1819(9-10):1080-7. doi: 10.1016/j.bbagrm.2011.11.008. Epub 2011 Dec 8. Biochim Biophys Acta. 2012. PMID: 22172992 Free PMC article. Review.
-
Effect of Lon protease knockdown on mitochondrial function in HeLa cells.Biochimie. 2014 May;100:38-47. doi: 10.1016/j.biochi.2013.12.005. Epub 2013 Dec 17. Biochimie. 2014. PMID: 24355201
-
Towards the control of intracellular protein turnover: mitochondrial Lon protease inhibitors versus proteasome inhibitors.Biochimie. 2008 Feb;90(2):260-9. doi: 10.1016/j.biochi.2007.10.010. Epub 2007 Oct 25. Biochimie. 2008. PMID: 18021745 Review.
Cited by
-
Whole genome expression profile in neuroblastoma cells exposed to 1-methyl-4-phenylpyridine.Neurotoxicology. 2012 Oct;33(5):1156-69. doi: 10.1016/j.neuro.2012.06.009. Epub 2012 Jul 7. Neurotoxicology. 2012. PMID: 22776087 Free PMC article.
-
Multitasking in the mitochondrion by the ATP-dependent Lon protease.Biochim Biophys Acta. 2012 Jan;1823(1):56-66. doi: 10.1016/j.bbamcr.2011.11.003. Epub 2011 Nov 18. Biochim Biophys Acta. 2012. PMID: 22119779 Free PMC article. Review.
-
Harnessing mitochondrial biogenesis to combat acute kidney injury: Current insights and futuredirections.Genes Dis. 2025 Apr 15;12(6):101645. doi: 10.1016/j.gendis.2025.101645. eCollection 2025 Nov. Genes Dis. 2025. PMID: 40821125 Free PMC article. Review.
-
Release of skeletal muscle peptide fragments identifies individual proteins degraded during insulin deprivation in type 1 diabetic humans and mice.Am J Physiol Endocrinol Metab. 2016 Sep 1;311(3):E628-37. doi: 10.1152/ajpendo.00175.2016. Epub 2016 Jul 19. Am J Physiol Endocrinol Metab. 2016. PMID: 27436610 Free PMC article.
-
CODAS syndrome is associated with mutations of LONP1, encoding mitochondrial AAA+ Lon protease.Am J Hum Genet. 2015 Jan 8;96(1):121-35. doi: 10.1016/j.ajhg.2014.12.003. Am J Hum Genet. 2015. PMID: 25574826 Free PMC article.
References
-
- Kang SG, Ortega J, Singh SK, Wang N, Huang NN, Steven AC, Maurizi MR. Functional proteolytic complexes of the human mitochondrial ATP-dependent protease, hClpXP. J Biol Chem. 2002;277:21095–21102. - PubMed
-
- Corydon TJ, Wilsbech M, Jespersgaard C, Andresen BS, Borglum AD, Pedersen S, Bolund L, Gregersen N, Bross P. Human and mouse mitochondrial orthologs of bacterial ClpX. Mamm Genome. 2000;11:899–905. - PubMed
-
- Bae MK, Jeong JW, Kim SH, Kim SY, Kang HJ, Kim DM, Bae SK, Yun I, Trentin GA, Rozakis-Adcock M, Kim KW. Tid-1 interacts with the von Hippel-Lindau protein and modulates angiogenesis by destabilization of HIF-1alpha. Cancer Res. 2005;65:2520–2525. - PubMed
-
- Im YJ, Na Y, Kang GB, Rho SH, Kim MK, Lee JH, Chung CH, Eom SH. The active site of a lon protease from Methanococcus jannaschii distinctly differs from the canonical catalytic Dyad of Lon proteases. J Biol Chem. 2004;279:53451–53457. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

