ScanRanker: Quality assessment of tandem mass spectra via sequence tagging
- PMID: 21520941
- PMCID: PMC3128668
- DOI: 10.1021/pr200118r
ScanRanker: Quality assessment of tandem mass spectra via sequence tagging
Abstract
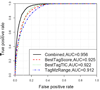
In shotgun proteomics, protein identification by tandem mass spectrometry relies on bioinformatics tools. Despite recent improvements in identification algorithms, a significant number of high quality spectra remain unidentified for various reasons. Here we present ScanRanker, an open-source tool that evaluates the quality of tandem mass spectra via sequence tagging with reliable performance in data from different instruments. The superior performance of ScanRanker enables it not only to find unassigned high quality spectra that evade identification through database search but also to select spectra for de novo sequencing and cross-linking analysis. In addition, we demonstrate that the distribution of ScanRanker scores predicts the richness of identifiable spectra among multiple LC-MS/MS runs in an experiment, and ScanRanker scores assist the process of peptide assignment validation to increase confident spectrum identifications. The source code and executable versions of ScanRanker are available from http://fenchurch.mc.vanderbilt.edu.
Figures





References
-
- Eng JKMAL, Yates JR. An approach to correlate tandem mass-spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994;5:976–989. - PubMed
-
- Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20(18):3551–3567. - PubMed
-
- Mann M, Wilm M. Error-tolerant identification of peptides in sequence databases by peptide sequence tags. Anal Chem. 1994;66(24):4390–4399. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

