Novel simple sequence repeats (SSRs) detected by ND-FISH in heterochromatin of Drosophila melanogaster
- PMID: 21521504
- PMCID: PMC3114746
- DOI: 10.1186/1471-2164-12-205
Novel simple sequence repeats (SSRs) detected by ND-FISH in heterochromatin of Drosophila melanogaster
Abstract
Background: In recent years, substantial progress has been made in understanding the organization of sequences in heterochromatin regions containing single-copy genes and transposable elements. However, the sequence and organization of tandem repeat DNA sequences, which are by far the majority fraction of D. melanogaster heterochromatin, are little understood.
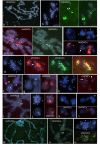
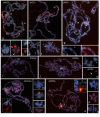
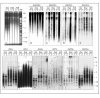
Results: This paper reports that the heterochromatin, as well as containing long tandem arrays of pentanucleotide satellites (AAGAG, AAGAC, AATAT, AATAC and AACAC), is also enriched in other simple sequence repeats (SSRs) such as A, AC, AG, AAG, ACT, GATA and GACA. Non-denaturing FISH (ND-FISH) showed these SSRs to localize to the chromocentre of polytene chromosomes, and was used to map them on mitotic chromosomes. Different distributions were detected ranging from single heterochromatic clusters to complex combinations on different chromosomes. ND-FISH performed on extended DNA fibres, along with Southern blotting, showed the complex organization of these heterochromatin sequences in long tracts, and revealed subclusters of SSRs (several kilobase in length) flanked by other DNA sequences. The chromosomal characterization of C, AAC, AGG, AAT, CCG, ACG, AGC, ATC and ACC provided further detailed information on the SSR content of D. melanogaster at the whole genome level.
Conclusion: These data clearly show the variation in the abundance of different SSR motifs and reveal their non-random distribution within and between chromosomes. The greater representation of certain SSRs in D. melanogaster heterochromatin suggests that its complexity may be greater than previously thought.
Figures



Similar articles
-
Physical organisation of simple sequence repeats (SSRs) in Triticeae: structural, functional and evolutionary implications.Cytogenet Genome Res. 2008;120(3-4):210-9. doi: 10.1159/000121069. Epub 2008 May 22. Cytogenet Genome Res. 2008. PMID: 18504349 Review.
-
Increasing the physical markers of wheat chromosomes using SSRs as FISH probes.Genome. 2008 Oct;51(10):809-15. doi: 10.1139/G08-065. Genome. 2008. PMID: 18923532
-
The nonrandom distribution of long clusters of all possible classes of trinucleotide repeats in barley chromosomes.Chromosome Res. 2007;15(6):711-20. doi: 10.1007/s10577-007-1156-8. Epub 2007 Aug 23. Chromosome Res. 2007. PMID: 17874212
-
Mapping simple repeated DNA sequences in heterochromatin of Drosophila melanogaster.Genetics. 1993 Aug;134(4):1149-74. doi: 10.1093/genetics/134.4.1149. Genetics. 1993. PMID: 8375654 Free PMC article.
-
Intercalary heterochromatin in polytene chromosomes of Drosophila melanogaster.Chromosoma. 2008 Oct;117(5):411-8. doi: 10.1007/s00412-008-0163-7. Epub 2008 May 20. Chromosoma. 2008. PMID: 18491121 Review.
Cited by
-
Chromosomal markers in the genus Karenia: Towards an understanding of the evolution of the chromosomes, life cycle patterns and phylogenetic relationships in dinoflagellates.Sci Rep. 2019 Feb 28;9(1):3072. doi: 10.1038/s41598-018-35785-7. Sci Rep. 2019. PMID: 30816125 Free PMC article.
-
Microsatellite organization in the grasshopper Abracris flavolineata (Orthoptera: Acrididae) revealed by FISH mapping: remarkable spreading in the A and B chromosomes.PLoS One. 2014 May 28;9(5):e97956. doi: 10.1371/journal.pone.0097956. eCollection 2014. PLoS One. 2014. PMID: 24871300 Free PMC article.
-
Hawaiian Drosophila genomes: size variation and evolutionary expansions.Genetica. 2016 Feb;144(1):107-24. doi: 10.1007/s10709-016-9882-5. Epub 2016 Jan 20. Genetica. 2016. PMID: 26790663
-
Physical location of SSR regions and cytogenetic instabilities in Pinus sylvestris chromosomes revealed by ND-FISH.J Genet. 2014 Aug;93(2):567-71. doi: 10.1007/s12041-014-0412-x. J Genet. 2014. PMID: 25189261 No abstract available.
-
Cytogenetic Diversity of Simple Sequences Repeats in Morphotypes of Brassica rapa ssp. chinensis.Front Plant Sci. 2016 Jul 26;7:1049. doi: 10.3389/fpls.2016.01049. eCollection 2016. Front Plant Sci. 2016. PMID: 27507974 Free PMC article.
References
-
- Grewal SI, Elgin SC. Heterochromatin: new possibilities for the inherence of structure. Curr Opin Genet Dev. 2002;12:178–187. - PubMed
-
- Morris CA, Moazed D. Centromere assembly and propagation. Cell. 2007;128(4):647–650. - PubMed
-
- Martienssen RA. Maintenance of heterochromatin by RNA interference of tandem repeats. Nat Genet. 2007;35(3):213–214. - PubMed
-
- Plohl M, Luchetti A, Mestrovic N, Mantovani B. Satellite DNAs between selfishness and functionality: Structure, genomics and evolution of tandem repeats in centromeric (hetero)chromatin. Gene. 2008;409:72–82. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

