Association studies for next-generation sequencing
- PMID: 21521787
- PMCID: PMC3129252
- DOI: 10.1101/gr.115998.110
Association studies for next-generation sequencing
Abstract
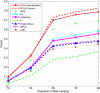
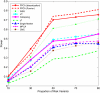
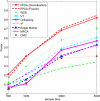
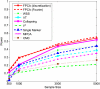
Genome-wide association studies (GWAS) have become the primary approach for identifying genes with common variants influencing complex diseases. Despite considerable progress, the common variations identified by GWAS account for only a small fraction of disease heritability and are unlikely to explain the majority of phenotypic variations of common diseases. A potential source of the missing heritability is the contribution of rare variants. Next-generation sequencing technologies will detect millions of novel rare variants, but these technologies have three defining features: identification of a large number of rare variants, a high proportion of sequence errors, and a large proportion of missing data. These features raise challenges for testing the association of rare variants with phenotypes of interest. In this study, we use a genome continuum model and functional principal components as a general principle for developing novel and powerful association analysis methods designed for resequencing data. We use simulations to calculate the type I error rates and the power of nine alternative statistics: two functional principal component analysis (FPCA)-based statistics, the multivariate principal component analysis (MPCA)-based statistic, the weighted sum (WSS), the variable-threshold (VT) method, the generalized T(2), the collapsing method, the CMC method, and individual tests. We also examined the impact of sequence errors on their type I error rates. Finally, we apply the nine statistics to the published resequencing data set from ANGPTL4 in the Dallas Heart Study. We report that FPCA-based statistics have a higher power to detect association of rare variants and a stronger ability to filter sequence errors than the other seven methods.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
