Applied genomics: data mining reveals species-specific malaria diagnostic targets more sensitive than 18S rRNA
- PMID: 21525225
- PMCID: PMC3147814
- DOI: 10.1128/JCM.02603-10
Applied genomics: data mining reveals species-specific malaria diagnostic targets more sensitive than 18S rRNA
Abstract
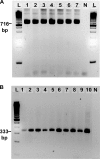
Accurate and rapid diagnosis of malaria infections is crucial for implementing species-appropriate treatment and saving lives. Molecular diagnostic tools are the most accurate and sensitive method of detecting Plasmodium, differentiating between Plasmodium species, and detecting subclinical infections. Despite available whole-genome sequence data for Plasmodium falciparum and P. vivax, the majority of PCR-based methods still rely on the 18S rRNA gene targets. Historically, this gene has served as the best target for diagnostic assays. However, it is limited in its ability to detect mixed infections in multiplex assay platforms without the use of nested PCR. New diagnostic targets are needed. Ideal targets will be species specific, highly sensitive, and amenable to both single-step and multiplex PCRs. We have mined the genomes of P. falciparum and P. vivax to identify species-specific, repetitive sequences that serve as new PCR targets for the detection of malaria. We show that these targets (Pvr47 and Pfr364) exist in 14 to 41 copies and are more sensitive than 18S rRNA when utilized in a single-step PCR. Parasites are routinely detected at levels of 1 to 10 parasites/μl. The reaction can be multiplexed to detect both species in a single reaction. We have examined 7 P. falciparum strains and 91 P. falciparum clinical isolates from Tanzania and 10 P. vivax strains and 96 P. vivax clinical isolates from Venezuela, and we have verified a sensitivity and specificity of ∼100% for both targets compared with a nested 18S rRNA approach. We show that bioinformatics approaches can be successfully applied to identify novel diagnostic targets and improve molecular methods for pathogen detection. These novel targets provide a powerful alternative molecular diagnostic method for the detection of P. falciparum and P. vivax in conventional or multiplex PCR platforms.
Figures




References
-
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 - PubMed
-
- Barcus M. J., et al. 2007. Demographic risk factors for severe and fatal vivax and falciparum malaria among hospital admissions in northeastern Indonesian Papua. Am. J. Trop. Med. Hyg. 77:984–991 - PubMed
-
- Bronzan R. N., McMorrow M. L., Kachur S. P. 2008. Diagnosis of malaria: challenges for clinicians in endemic and non-endemic regions. Mol. Diagn. Ther. 12:299–306 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

