RAB11A is essential for transport of the influenza virus genome to the plasma membrane
- PMID: 21525351
- PMCID: PMC3126513
- DOI: 10.1128/JVI.00378-11
RAB11A is essential for transport of the influenza virus genome to the plasma membrane
Abstract
Influenza A virus assembly is a complex process that requires the intersection of pathways involved in transporting viral glycoproteins, the matrix protein, and viral genomes, incorporated in the viral ribonucleoprotein (vRNP) complex, to plasma membrane sites of virion formation. Among these virion components, the mechanism of vRNP delivery is the most incompletely understood. Here, we reveal a functional relationship between the cellular Rab11 GTPase isoform, RAB11A, and vRNPs and show that RAB11A is indispensable for proper vRNP transport to the plasma membrane. Using an immunofluorescence-based assay with a monoclonal antibody that recognizes nucleoprotein in the form of vRNP, we demonstrate association between RAB11A and vRNPs at all stages of vRNP cytoplasmic transport. Abrogation of RAB11A expression through small interfering RNA (siRNA) treatment or disruption of RAB11A function by overexpression of dominant negative or constitutively active proteins caused aberrant vRNP intracellular accumulation, retention in the perinuclear region, and lack of accumulation at the plasma membrane. Complex formation between RAB11A and vRNPs was further established biochemically. Our results uncover a critical host factor with an essential contribution to influenza virus genome delivery and reveal a potential role for RAB11A in the transport of ribonucleoprotein cargo.
Figures
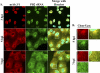
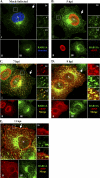
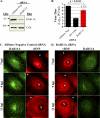
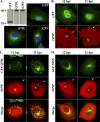


References
-
- Amorim M. J., et al. 9 February 2011. A Rab11 and microtubule dependent mechanism for cytoplasmic transport of influenza A virus vRNA. J. Virol. doi:10.1128/JVI.02606-10 - DOI - PMC - PubMed
-
- Arcangeletti M. C., et al. 1997. Modification of cytoskeleton and prosome networks in relation to protein synthesis in influenza A virus-infected LLC-MK2 cells. Virus Res. 51:19–34 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

