Gemcitabine/cannabinoid combination triggers autophagy in pancreatic cancer cells through a ROS-mediated mechanism
- PMID: 21525939
- PMCID: PMC3122066
- DOI: 10.1038/cddis.2011.36
Gemcitabine/cannabinoid combination triggers autophagy in pancreatic cancer cells through a ROS-mediated mechanism
Abstract
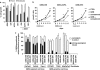
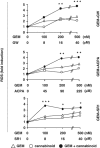
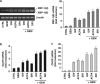
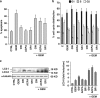
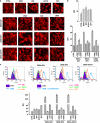
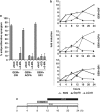
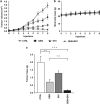
Gemcitabine (GEM, 2',2'-difluorodeoxycytidine) is currently used in advanced pancreatic adenocarcinoma, with a response rate of < 20%. The purpose of our work was to improve GEM activity by addition of cannabinoids. Here, we show that GEM induces both cannabinoid receptor-1 (CB1) and cannabinoid receptor-2 (CB2) receptors by an NF-κB-dependent mechanism and that its association with cannabinoids synergistically inhibits pancreatic adenocarcinoma cell growth and increases reactive oxygen species (ROS) induced by single treatments. The antiproliferative synergism is prevented by the radical scavenger N-acetyl-L-cysteine and by the specific NF-κB inhibitor BAY 11-7085, demonstrating that the induction of ROS by GEM/cannabinoids and of NF-κB by GEM is required for this effect. In addition, we report that neither apoptotic nor cytostatic mechanisms are responsible for the synergistic cell growth inhibition, which is strictly associated with the enhancement of endoplasmic reticulum stress and autophagic cell death. Noteworthy, the antiproliferative synergism is stronger in GEM-resistant pancreatic cancer cell lines compared with GEM-sensitive pancreatic cancer cell lines. The combined treatment strongly inhibits growth of human pancreatic tumor cells xenografted in nude mice without apparent toxic effects. These findings support a key role of the ROS-dependent activation of an autophagic program in the synergistic growth inhibition induced by GEM/cannabinoid combination in human pancreatic cancer cells.
Figures

References
-
- Li J, Merl MY, Chabot J, Saif MW. Updates of adjuvant therapy in pancreatic cancer: where are we and where are we going? Highlights from the ‘2010 ASCO Annual Meeting'. Chicago, IL, USA. JOP. 2010;11:310–312. - PubMed
-
- Neoptolemos JP, Cunningham D, Friess H, Bassi C, Stocken DD, Tait DM, et al. Adjuvant therapy in pancreatic cancer: historical and current perspectives. Ann Oncol. 2003;14:675–692. - PubMed
-
- Burris HA, III, Moore MJ, Andersen J, Green MR, Rothenberg ML, Modiano MR, et al. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: a randomized trial. J Clin Oncol. 1997;15:2403–2413. - PubMed
-
- Laurent A, Nicco C, Chereau C, Goulvestre C, Alexandre J, Alves A, et al. Controlling tumor growth by modulating endogenous production of reactive oxygen species. Cancer Res. 2005;65:948–956. - PubMed
-
- Donadelli M, Costanzo C, Beghelli S, Scupoli MT, Dandrea M, Bonora A, et al. Synergistic inhibition of pancreatic adenocarcinoma cell growth by trichostatin A and gemcitabine. Biochim Biophys Acta. 2007;1773:1095–1106. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

