The telomere binding protein TRF2 induces chromatin compaction
- PMID: 21526145
- PMCID: PMC3079743
- DOI: 10.1371/journal.pone.0019124
The telomere binding protein TRF2 induces chromatin compaction
Abstract
Mammalian telomeres are specialized chromatin structures that require the telomere binding protein, TRF2, for maintaining chromosome stability. In addition to its ability to modulate DNA repair activities, TRF2 also has direct effects on DNA structure and topology. Given that mammalian telomeric chromatin includes nucleosomes, we investigated the effect of this protein on chromatin structure. TRF2 bound to reconstituted telomeric nucleosomal fibers through both its basic N-terminus and its C-terminal DNA binding domain. Analytical agarose gel electrophoresis (AAGE) studies showed that TRF2 promoted the folding of nucleosomal arrays into more compact structures by neutralizing negative surface charge. A construct containing the N-terminal and TRFH domains together altered the charge and radius of nucleosomal arrays similarly to full-length TRF2 suggesting that TRF2-driven changes in global chromatin structure were largely due to these regions. However, the most compact chromatin structures were induced by the isolated basic N-terminal region, as judged by both AAGE and atomic force microscopy. Although the N-terminal region condensed nucleosomal array fibers, the TRFH domain, known to alter DNA topology, was required for stimulation of a strand invasion-like reaction with nucleosomal arrays. Optimal strand invasion also required the C-terminal DNA binding domain. Furthermore, the reaction was not stimulated on linear histone-free DNA. Our data suggest that nucleosomal chromatin has the ability to facilitate this activity of TRF2 which is thought to be involved in stabilizing looped telomere structures.
Conflict of interest statement
Figures




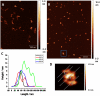
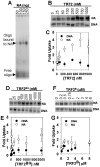
References
-
- Falk M, Lukasova E, Kozubek S. Higher-order chromatin structure in DSB induction, repair and misrepair. Mutat Res. 2010;704:88–100. - PubMed
-
- Hansen JC. Conformational dynamics of the chromatin fiber in solution: determinants, mechanisms, and functions. Annu Rev Biophys Biomol Struct. 2002;31:361–392. - PubMed
-
- Horn PJ, Peterson CL. Molecular biology. Chromatin higher order folding—wrapping up transcription. Science. 2002;297:1824–1827. - PubMed
-
- Luger K, Hansen JC. Nucleosome and chromatin fiber dynamics. Curr Opin Struct Biol. 2005;15:188–196. - PubMed
-
- Lu X, Klonoski JM, Resch MG, Hansen JC. In vitro chromatin self-association and its relevance to genome architecture. Biochem Cell Biol. 2006;84:411–417. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

