Deciphering the role of RND efflux transporters in Burkholderia cenocepacia
- PMID: 21526150
- PMCID: PMC3079749
- DOI: 10.1371/journal.pone.0018902
Deciphering the role of RND efflux transporters in Burkholderia cenocepacia
Abstract
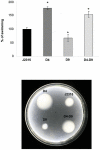
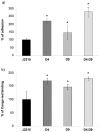
Burkholderia cenocepacia J2315 is representative of a highly problematic group of cystic fibrosis (CF) pathogens. Eradication of B. cenocepacia is very difficult with the antimicrobial therapy being ineffective due to its high resistance to clinically relevant antimicrobial agents and disinfectants. RND (Resistance-Nodulation-Cell Division) efflux pumps are known to be among the mediators of multidrug resistance in gram-negative bacteria. Since the significance of the 16 RND efflux systems present in B. cenocepacia (named RND-1 to -16) has been only partially determined, the aim of this work was to analyze mutants of B. cenocepacia strain J2315 impaired in RND-4 and RND-9 efflux systems, and assess their role in the efflux of toxic compounds. The transcriptomes of mutants deleted individually in RND-4 and RND-9 (named D4 and D9), and a double-mutant in both efflux pumps (named D4-D9), were compared to that of the wild-type B. cenocepacia using microarray analysis. Microarray data were confirmed by qRT-PCR, phenotypic experiments, and by Phenotype MicroArray analysis. The data revealed that RND-4 made a significant contribution to the antibiotic resistance of B. cenocepacia, whereas RND-9 was only marginally involved in this process. Moreover, the double mutant D4-D9 showed a phenotype and an expression profile similar to D4. The microarray data showed that motility and chemotaxis-related genes appeared to be up-regulated in both D4 and D4-D9 strains. In contrast, these gene sets were down-regulated or expressed at levels similar to J2315 in the D9 mutant. Biofilm production was enhanced in all mutants. Overall, these results indicate that in B. cenocepacia RND pumps play a wider role than just in drug resistance, influencing additional phenotypic traits important for pathogenesis.
Conflict of interest statement
Figures

References
-
- Mahenthiralingam E, Baldwin A, Dowson CG. Burkholderia cenocepacia complex bacteria: opportunistic pathogens with important natural biology. J Appl Microbiol. 2008;104:1539–1551. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

