The complete genome sequence of Fibrobacter succinogenes S85 reveals a cellulolytic and metabolic specialist
- PMID: 21526192
- PMCID: PMC3079729
- DOI: 10.1371/journal.pone.0018814
The complete genome sequence of Fibrobacter succinogenes S85 reveals a cellulolytic and metabolic specialist
Abstract
Fibrobacter succinogenes is an important member of the rumen microbial community that converts plant biomass into nutrients usable by its host. This bacterium, which is also one of only two cultivated species in its phylum, is an efficient and prolific degrader of cellulose. Specifically, it has a particularly high activity against crystalline cellulose that requires close physical contact with this substrate. However, unlike other known cellulolytic microbes, it does not degrade cellulose using a cellulosome or by producing high extracellular titers of cellulase enzymes. To better understand the biology of F. succinogenes, we sequenced the genome of the type strain S85 to completion. A total of 3,085 open reading frames were predicted from its 3.84 Mbp genome. Analysis of sequences predicted to encode for carbohydrate-degrading enzymes revealed an unusually high number of genes that were classified into 49 different families of glycoside hydrolases, carbohydrate binding modules (CBMs), carbohydrate esterases, and polysaccharide lyases. Of the 31 identified cellulases, none contain CBMs in families 1, 2, and 3, typically associated with crystalline cellulose degradation. Polysaccharide hydrolysis and utilization assays showed that F. succinogenes was able to hydrolyze a number of polysaccharides, but could only utilize the hydrolytic products of cellulose. This suggests that F. succinogenes uses its array of hemicellulose-degrading enzymes to remove hemicelluloses to gain access to cellulose. This is reflected in its genome, as F. succinogenes lacks many of the genes necessary to transport and metabolize the hydrolytic products of non-cellulose polysaccharides. The F. succinogenes genome reveals a bacterium that specializes in cellulose as its sole energy source, and provides insight into a novel strategy for cellulose degradation.
Conflict of interest statement
Figures
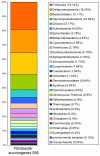
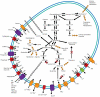
References
-
- Russell JB. Ithaca, NY: J.B. Russell Publishing Co; 2002. Rumen microbiology and its role in ruminant nutrition.
-
- Flint HJ, Bayer EA, Rincon MT, Lamed R, White BA. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat Rev Micro. 2008;6:121–131. - PubMed
-
- Montgomery L, Flesher B, Stahl D. Transfer of Bacteroides succinogenes (Hungate) to Fibrobacter gen. nov. as Fibrobacter succinogenes comb. nov. and Description of Fibrobacter intestinalis sp. nov. Int J Syst Evol Microbiol. 1988;38:430–435.
-
- Stevenson DM, Weimer PJ. Dominance of Prevotella and low abundance of classical ruminal bacterial species in the bovine rumen revealed by relative quantification real-time PCR. Appl Microbiol Biotechnol. 2007;75:165–174. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

