The analysis of para-cresol production and tolerance in Clostridium difficile 027 and 012 strains
- PMID: 21527013
- PMCID: PMC3102038
- DOI: 10.1186/1471-2180-11-86
The analysis of para-cresol production and tolerance in Clostridium difficile 027 and 012 strains
Abstract
Background: Clostridium difficile is the major cause of antibiotic associated diarrhoea and in recent years its increased prevalence has been linked to the emergence of hypervirulent clones such as the PCR-ribotype 027. Characteristically, C. difficile infection (CDI) occurs after treatment with broad-spectrum antibiotics, which disrupt the normal gut microflora and allow C. difficile to flourish. One of the relatively unique features of C. difficile is its ability to ferment tyrosine to para-cresol via the intermediate para-hydroxyphenylacetate (p-HPA). P-cresol is a phenolic compound with bacteriostatic properties which C. difficile can tolerate and may provide the organism with a competitive advantage over other gut microflora, enabling it to proliferate and cause CDI. It has been proposed that the hpdBCA operon, rarely found in other gut microflora, encodes the enzymes responsible for the conversion of p-HPA to p-cresol.
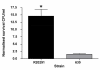
Results: We show that the PCR-ribotype 027 strain R20291 quantitatively produced more p-cresol in-vitro and was significantly more tolerant to p-cresol than the sequenced strain 630 (PCR-ribotype 012). Tyrosine conversion to p-HPA was only observed under certain conditions. We constructed gene inactivation mutants in the hpdBCA operon in strains R20291 and 630Δerm which curtails their ability to produce p-cresol, confirming the role of these genes in p-cresol production. The mutants were equally able to tolerate p-cresol compared to the respective parent strains, suggesting that tolerance to p-cresol is not linked to its production.
Conclusions: C. difficile converts tyrosine to p-cresol, utilising the hpdBCA operon in C. difficile strains 630 and R20291. The hypervirulent strain R20291 exhibits increased production of and tolerance to p-cresol, which may be a contributory factor to the virulence of this strain and other hypervirulent PCR-ribotype 027 strains.
Figures




Similar articles
-
Clostridioides difficile para-Cresol Production Is Induced by the Precursor para-Hydroxyphenylacetate.J Bacteriol. 2020 Aug 25;202(18):e00282-20. doi: 10.1128/JB.00282-20. Print 2020 Aug 25. J Bacteriol. 2020. PMID: 32631945 Free PMC article.
-
Production of p-cresol by Decarboxylation of p-HPA by All Five Lineages of Clostridioides difficile Provides a Growth Advantage.Front Cell Infect Microbiol. 2021 Oct 29;11:757599. doi: 10.3389/fcimb.2021.757599. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34778108 Free PMC article.
-
Para-cresol production by Clostridium difficile affects microbial diversity and membrane integrity of Gram-negative bacteria.PLoS Pathog. 2018 Sep 12;14(9):e1007191. doi: 10.1371/journal.ppat.1007191. eCollection 2018 Sep. PLoS Pathog. 2018. PMID: 30208103 Free PMC article.
-
Regulation of para-cresol production in Clostridioides difficile.Curr Opin Microbiol. 2022 Feb;65:131-137. doi: 10.1016/j.mib.2021.11.005. Epub 2021 Nov 29. Curr Opin Microbiol. 2022. PMID: 34856509 Review.
-
Clostridium difficile infection: molecular pathogenesis and novel therapeutics.Expert Rev Anti Infect Ther. 2014 Jan;12(1):131-50. doi: 10.1586/14787210.2014.866515. Expert Rev Anti Infect Ther. 2014. PMID: 24410618 Free PMC article. Review.
Cited by
-
Epidemic Clostridium difficile ribotype 027 in Chile.Emerg Infect Dis. 2012 Aug;18(8):1370-2. doi: 10.3201/eid1808.120211. Emerg Infect Dis. 2012. PMID: 22840230 Free PMC article. No abstract available.
-
Microbiota-gut-brain axis and ketogenic diet: how close are we to tackling epilepsy?Microbiome Res Rep. 2023 Aug 29;2(4):32. doi: 10.20517/mrr.2023.24. eCollection 2023. Microbiome Res Rep. 2023. PMID: 38045924 Free PMC article. Review.
-
Environmental Chemical Diethylhexyl Phthalate Alters Intestinal Microbiota Community Structure and Metabolite Profile in Mice.mSystems. 2019 Dec 10;4(6):e00724-19. doi: 10.1128/mSystems.00724-19. mSystems. 2019. PMID: 31822602 Free PMC article.
-
Interspecies Interactions between Clostridium difficile and Candida albicans.mSphere. 2016 Nov 9;1(6):e00187-16. doi: 10.1128/mSphere.00187-16. eCollection 2016 Nov-Dec. mSphere. 2016. PMID: 27840850 Free PMC article.
-
Plasma microbiome-modulated indole- and phenyl-derived metabolites associate with advanced atherosclerosis and postoperative outcomes.J Vasc Surg. 2018 Nov;68(5):1552-1562.e7. doi: 10.1016/j.jvs.2017.09.029. Epub 2017 Dec 13. J Vasc Surg. 2018. PMID: 29248242 Free PMC article.
References
-
- Brazier JS, Raybould R, Patel B, Duckworth G, Pearson A, Charlett A, Duerden BI. Distribution and antimicrobial susceptibility patterns of Clostridium difficile PCR ribotypes in English hospitals, 2007-08. Euro Surveill. 2008;13(41) - PubMed
-
- Loo VG, Poirier L, Miller MA, Oughton M, Libman MD, Michaud S, Bourgault AM, Nguyen T, Frenette C, Kelly M, Vibien A, Brassard P, Fenn S, Dewar K, Hudson TJ, Horn R, Rene P, Monczak Y, Dascal A. A predominantly clonal multi-institutional outbreak of Clostridium difficile-associated diarrhea with high morbidity and mortality. N Engl J Med. 2005;353(23):2442–2449. doi: 10.1056/NEJMoa051639. - DOI - PubMed
-
- Goorhuis A, Van der Kooi T, Vaessen N, Dekker FW, Van den Berg R, Harmanus C, van den Hof S, Notermans DW, Kuijper EJ. Spread and epidemiology of Clostridium difficile polymerase chain reaction ribotype 027/toxinotype III in the Netherlands. Clin Infect Dis. 2007;45(6):695–703. doi: 10.1086/520984. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

