Methods for the Analysis of High Precision Differential Hydrogen Deuterium Exchange Data
- PMID: 21528013
- PMCID: PMC3081588
- DOI: 10.1016/j.ijms.2010.08.002
Methods for the Analysis of High Precision Differential Hydrogen Deuterium Exchange Data
Abstract
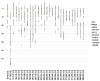
Hydrogen/deuterium exchange (HDX) mass spectrometry has been widely applied to the characterization of protein dynamics. More recently, differential HDX has been shown to be effective for the characterization of ligand binding. Previously we have described a fully automated HDX system for use as a ligand screening platform. Here we describe and validate the required data analysis workflow to facilitate the use of HDX as a robust approach for ligand screening. Following acquisition of HDX data at a single on-exchange time point (n ≥ 3), one way analysis of variance in conjunction with the Tukey multiple comparison procedure is used to establish the significance of any measured difference. Analysis results are graphed with respect to a single peptide, ligand or group of ligands, or displayed as an overview within a heat map. For the heat map display, only Δ%D values with a Tukey-adjusted P value less than 0.05 are colored. Hierarchical clustering is used to bin compounds with highly similar HDX signatures. The workflow is evaluated with a small data set showing the ligand binding domain (LDB) of the nuclear receptor peroxisome proliferator-activated receptor gamma (PPARγ) screened against 10 functionally selective ligands. More significantly, data for the vitamin D receptor (VDR) in complex with 87 ligands are presented. To highlight the robustness and precision of our automated HDX platform we analyzed the data from 4191 replicate HDX measurements acquired over an eight month timeframe. Ninety six percent of these measurements were within 10 percent of the mean value. Work has begun to integrate these analysis and graphing components within our HDX software suite.
Figures

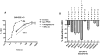
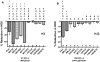



References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
