Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes
- PMID: 21529714
- PMCID: PMC3085131
- DOI: 10.1016/j.cell.2011.03.031
Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes
Abstract
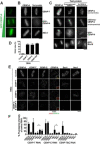
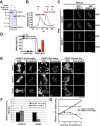
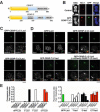
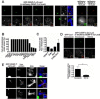
Accurate chromosome segregation requires assembly of the multiprotein kinetochore complex at centromeres. Although prior work identified the centromeric histone H3-variant CENP-A as the important upstream factor necessary for centromere specification, in human cells CENP-A is not sufficient for kinetochore assembly. Here, we demonstrate that two constitutive DNA-binding kinetochore components, CENP-C and CENP-T, function to direct kinetochore formation. Replacing the DNA-binding regions of CENP-C and CENP-T with alternate chromosome-targeting domains recruits these proteins to ectopic loci, resulting in CENP-A-independent kinetochore assembly. These ectopic kinetochore-like foci are functional based on the stoichiometric assembly of multiple kinetochore components, including the microtubule-binding KMN network, the presence of microtubule attachments, the microtubule-sensitive recruitment of the spindle checkpoint protein Mad2, and the segregation behavior of foci-containing chromosomes. We additionally find that CENP-T phosphorylation regulates the mitotic assembly of both endogenous and ectopic kinetochores. Thus, CENP-C and CENP-T form a critical regulated platform for vertebrate kinetochore assembly.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures



References
-
- Cheeseman IM. A Combined Approach for the Localization and Tandem Affinity Purification of Protein Complexes from Metazoans. Science's STKE. 2005;2005:pl1–pl1. - PubMed
-
- Cheeseman IM, Desai A. Molecular architecture of the kinetochore- microtubule interface. Nat Rev Mol Cell Biol. 2008;9:33–46. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

