An epigenetic signature for monoallelic olfactory receptor expression
- PMID: 21529909
- PMCID: PMC3094500
- DOI: 10.1016/j.cell.2011.03.040
An epigenetic signature for monoallelic olfactory receptor expression
Abstract
Constitutive heterochromatin is traditionally viewed as the static form of heterochromatin that silences pericentromeric and telomeric repeats in a cell cycle- and differentiation-independent manner. Here, we show that, in the mouse olfactory epithelium, olfactory receptor (OR) genes are marked in a highly dynamic fashion with the molecular hallmarks of constitutive heterochromatin, H3K9me3 and H4K20me3. The cell type and developmentally dependent deposition of these marks along the OR clusters are, most likely, reversed during the process of OR choice to allow for monogenic and monoallelic OR expression. In contrast to the current view of OR choice, our data suggest that OR silencing takes place before OR expression, indicating that it is not the product of an OR-elicited feedback signal. Our findings suggest that chromatin-mediated silencing lays a molecular foundation upon which singular and stochastic selection for gene expression can be applied.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures

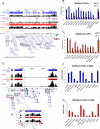
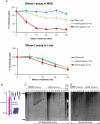
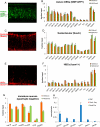
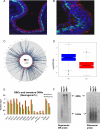

References
-
- Axel R. Scents and sensibility: a molecular logic of olfactory perception (Nobel lecture). Angewandte Chemie (International ed. 2005;44:6110–6127. - PubMed
-
- Barnea G, O'Donnell S, Mancia F, Sun X, Nemes A, Mendelsohn M, Axel R. Odorant receptors on axon termini in the brain. Science (New York, NY. 2004;304:1468. - PubMed
-
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. - PubMed
-
- Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–187. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

