CRISPR-based adaptive immune systems
- PMID: 21531607
- PMCID: PMC3119747
- DOI: 10.1016/j.mib.2011.03.005
CRISPR-based adaptive immune systems
Abstract
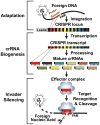
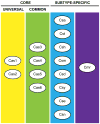
CRISPR-Cas systems are recently discovered, RNA-based immune systems that control invasions of viruses and plasmids in archaea and bacteria. Prokaryotes with CRISPR-Cas immune systems capture short invader sequences within the CRISPR loci in their genomes, and small RNAs produced from the CRISPR loci (CRISPR (cr)RNAs) guide Cas proteins to recognize and degrade (or otherwise silence) the invading nucleic acids. There are multiple variations of the pathway found among prokaryotes, each mediated by largely distinct components and mechanisms that we are only beginning to delineate. Here we will review our current understanding of the remarkable CRISPR-Cas pathways with particular attention to studies relevant to systems found in the archaea.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures

References
-
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

