Bi-specific aptamers mediating tumor cell lysis
- PMID: 21531729
- PMCID: PMC3122244
- DOI: 10.1074/jbc.M111.238261
Bi-specific aptamers mediating tumor cell lysis
Abstract
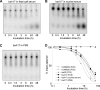
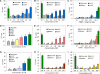
Antibody-dependent cellular cytotoxicity plays a pivotal role in antibody-based tumor therapies and is based on the recruitment of natural killer cells to antibody-bound tumor cells via binding of the Fcγ receptor III (CD16). Here we describe the generation of chimeric DNA aptamers that simultaneously bind to CD16α and c-Met, a receptor that is overexpressed in many tumors. By application of the systematic evolution of ligands by exponential enrichment (SELEX) method, CD16α specific DNA aptamers were isolated that bound with high specificity and affinity (91 pm-195 nm) to their respective recombinant and cellularly expressed target proteins. Two optimized CD16α specific aptamers were coupled to each of two c-Met specific aptamers using different linkers. Bi-specific aptamers retained suitable binding properties and displayed simultaneous binding to both antigens. Moreover, they mediated cellular cytotoxicity dependent on aptamer and effector cell concentration. Displacement of a bi-specific aptamer from CD16α by competing antibody 3G8 reduced cytotoxicity and confirmed the proposed mode of action. These results represent the first gain of a tumor-effective function of two distinct oligonucleotides by linkage into a bi-specific aptamer mediating cellular cytotoxicity.
Figures


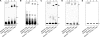
Similar articles
-
Absolute quantification of cell-bound DNA aptamers during SELEX.Nucleic Acid Ther. 2013 Apr;23(2):125-30. doi: 10.1089/nat.2012.0406. Epub 2013 Feb 13. Nucleic Acid Ther. 2013. PMID: 23405949
-
SELEX Modifications and Bioanalytical Techniques for Aptamer-Target Binding Characterization.Crit Rev Anal Chem. 2016 Nov;46(6):521-37. doi: 10.1080/10408347.2016.1157014. Epub 2016 Mar 15. Crit Rev Anal Chem. 2016. PMID: 26980177 Review.
-
Capillary electrophoresis-systematic evolution of ligands by exponential enrichment selection of base- and sugar-modified DNA aptamers: target binding dominated by 2'-O,4'-C-methylene-bridged/locked nucleic acid primer.Anal Chem. 2013 May 21;85(10):4961-7. doi: 10.1021/ac400058z. Epub 2013 May 10. Anal Chem. 2013. PMID: 23662585
-
DNA aptamers that bind to PrP(C) and not PrP(Sc) show sequence and structure specificity.Exp Biol Med (Maywood). 2006 Feb;231(2):204-14. doi: 10.1177/153537020623100211. Exp Biol Med (Maywood). 2006. PMID: 16446497
-
Post-SELEX optimization of aptamers.Anal Bioanal Chem. 2016 Jul;408(17):4567-73. doi: 10.1007/s00216-016-9556-2. Epub 2016 May 12. Anal Bioanal Chem. 2016. PMID: 27173394 Review.
Cited by
-
Recent advances in aptamer applications for analytical biochemistry.Anal Biochem. 2022 May 1;644:113894. doi: 10.1016/j.ab.2020.113894. Epub 2020 Aug 5. Anal Biochem. 2022. PMID: 32763306 Free PMC article.
-
RNA Aptamers Targeting Integrin α5β1 as Probes for Cyto- and Histofluorescence in Glioblastoma.Mol Ther Nucleic Acids. 2019 Sep 6;17:63-77. doi: 10.1016/j.omtn.2019.05.006. Epub 2019 May 24. Mol Ther Nucleic Acids. 2019. PMID: 31226519 Free PMC article.
-
Development of Cell-SELEX Technology and Its Application in Cancer Diagnosis and Therapy.Int J Mol Sci. 2016 Dec 10;17(12):2079. doi: 10.3390/ijms17122079. Int J Mol Sci. 2016. PMID: 27973403 Free PMC article. Review.
-
The Potential of Aptamer-Mediated Liquid Biopsy for Early Detection of Cancer.Int J Mol Sci. 2021 May 25;22(11):5601. doi: 10.3390/ijms22115601. Int J Mol Sci. 2021. PMID: 34070509 Free PMC article. Review.
-
Aptamers: A Feasible Technology in Cancer Immunotherapy.J Immunol Res. 2016;2016:1083738. doi: 10.1155/2016/1083738. Epub 2016 Jun 20. J Immunol Res. 2016. PMID: 27413756 Free PMC article. Review.
References
-
- Ruckman J., Green L. S., Beeson J., Waugh S., Gillette W. L., Henninger D. D., Claesson-Welsh L., Janjić N. (1998) J. Biol. Chem. 273, 20556–20567 - PubMed
-
- Ellington A. D., Szostak J. W. (1990) Nature 346, 818–822 - PubMed
-
- Tuerk C., Gold L. (1990) Science 249, 505–510 - PubMed
-
- Di Giusto D. A., Knox S. M., Lai Y., Tyrelle G. D., Aung M. T., King G. C. (2006) Chembiochem 7, 535–544 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

