Formation of a stable RuvA protein double tetramer is required for efficient branch migration in vitro and for replication fork reversal in vivo
- PMID: 21531731
- PMCID: PMC3121385
- DOI: 10.1074/jbc.M111.233908
Formation of a stable RuvA protein double tetramer is required for efficient branch migration in vitro and for replication fork reversal in vivo
Abstract
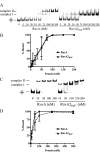
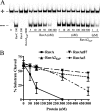
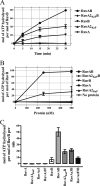
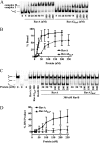
In bacteria, RuvABC is required for the resolution of Holliday junctions (HJ) made during homologous recombination. The RuvAB complex catalyzes HJ branch migration and replication fork reversal (RFR). During RFR, a stalled fork is reversed to form a HJ adjacent to a DNA double strand end, a reaction that requires RuvAB in certain Escherichia coli replication mutants. The exact structure of active RuvAB complexes remains elusive as it is still unknown whether one or two tetramers of RuvA support RuvB during branch migration and during RFR. We designed an E. coli RuvA mutant, RuvA2(KaP), specifically impaired for RuvA tetramer-tetramer interactions. As expected, the mutant protein is impaired for complex II (two tetramers) formation on HJs, although the binding efficiency of complex I (a single tetramer) is as wild type. We show that although RuvA complex II formation is required for efficient HJ branch migration in vitro, RuvA2(KaP) is fully active for homologous recombination in vivo. RuvA2(KaP) is also deficient at forming complex II on synthetic replication forks, and the binding affinity of RuvA2(KaP) for forks is decreased compared with wild type. Accordingly, RuvA2(KaP) is inefficient at processing forks in vitro and in vivo. These data indicate that RuvA2(KaP) is a separation-of-function mutant, capable of homologous recombination but impaired for RFR. RuvA2(KaP) is defective for stimulation of RuvB activity and stability of HJ·RuvA·RuvB tripartite complexes. This work demonstrates that the need for RuvA tetramer-tetramer interactions for full RuvAB activity in vitro causes specifically an RFR defect in vivo.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

