Aberrant epigenetic and genetic marks are seen in myelodysplastic leukocytes and reveal Dock4 as a candidate pathogenic gene on chromosome 7q
- PMID: 21532034
- PMCID: PMC3137092
- DOI: 10.1074/jbc.M111.235028
Aberrant epigenetic and genetic marks are seen in myelodysplastic leukocytes and reveal Dock4 as a candidate pathogenic gene on chromosome 7q
Abstract
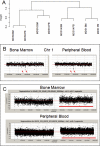
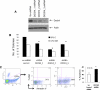
Myelodysplastic syndromes (MDS) are characterized by abnormal and dysplastic maturation of all blood lineages. Even though epigenetic alterations have been seen in MDS marrow progenitors, very little is known about the molecular alterations in dysplastic peripheral blood cells. We analyzed the methylome of MDS leukocytes by the HELP assay and determined that it was globally distinct from age-matched controls and was characterized by numerous novel, aberrant hypermethylated marks that were located mainly outside of CpG islands and preferentially affected GTPase regulators and other cancer-related pathways. Additionally, array comparative genomic hybridization revealed that novel as well as previously characterized deletions and amplifications could also be visualized in peripheral blood leukocytes, thus potentially reducing the need for bone marrow samples for future studies. Using integrative analysis, potentially pathogenic genes silenced by genetic deletions and aberrant hypermethylation in different patients were identified. DOCK4, a GTPase regulator located in the commonly deleted 7q31 region, was identified by this unbiased approach. Significant hypermethylation and reduced expression of DOCK4 in MDS bone marrow stem cells was observed in two large independent datasets, providing further validation of our findings. Finally, DOCK4 knockdown in primary marrow CD34(+) stem cells led to decreased erythroid colony formation and increased apoptosis, thus recapitulating the bone marrow failure seen in MDS. These findings reveal widespread novel epigenetic alterations in myelodysplastic leukocytes and implicate DOCK4 as a pathogenic gene located on the 7q chromosomal region.
Figures




References
-
- Pellagatti A., Cazzola M., Giagounidis A. A., Malcovati L., Porta M. G., Killick S., Campbell L. J., Wang L., Langford C. F., Fidler C., Oscier D., Aul C., Wainscoat J. S., Boultwood J. (2006) Blood 108, 337–345 - PubMed
-
- Jones P. A., Baylin S. B. (2002) Nat. Rev. Genet. 3, 415–428 - PubMed
-
- Christiansen D. H., Andersen M. K., Pedersen-Bjergaard J. (2003) Leukemia 17, 1813–1819 - PubMed
-
- Aggerholm A., Holm M. S., Guldberg P., Olesen L. H., Hokland P. (2006) Eur. J. Haematol. 76, 23–32 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

