Harnessing synthetic lethal interactions in anticancer drug discovery
- PMID: 21532565
- PMCID: PMC3652585
- DOI: 10.1038/nrd3374
Harnessing synthetic lethal interactions in anticancer drug discovery
Abstract
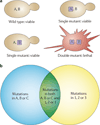
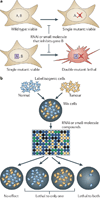
Unique features of tumours that can be exploited by targeted therapies are a key focus of current cancer research. One such approach is known as synthetic lethality screening, which involves searching for genetic interactions of two mutations whereby the presence of either mutation alone has no effect on cell viability but the combination of the two mutations results in cell death. The presence of one of these mutations in cancer cells but not in normal cells can therefore create opportunities to selectively kill cancer cells by mimicking the effect of the second genetic mutation with targeted therapy. Here, we summarize strategies that can be used to identify synthetic lethal interactions for anticancer drug discovery, describe examples of such interactions that are currently being investigated in preclinical and clinical studies of targeted anticancer therapies, and discuss the challenges of realizing the full potential of such therapies.
Figures

References
-
- Druker BJ. Perspectives on the development of a molecularly targeted agent. Cancer Cell. 2002;1:31–36. - PubMed
-
- Hellman S, Vokes EE. Advancing current treatments for cancer. Sci. Am. 1996;275:118–123. - PubMed
-
- Oliff A, Gibbs JB, McCormick F. New molecular targets for cancer therapy. Sci. Am. 1996;275:144–149. - PubMed
-
- Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nature Rev. Cancer. 2005;5:341–354. - PubMed
-
- Buchdunger E, et al. Inhibition of the Abl protein-tyrosine kinase in vitro and in vivo by a 2-phenylaminopyrimidine derivative. Cancer Res. 1996;56:100–104. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

