General properties of transcriptional time series in Escherichia coli
- PMID: 21532574
- PMCID: PMC3102781
- DOI: 10.1038/ng.821
General properties of transcriptional time series in Escherichia coli
Abstract
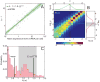
Gene activity is described by the time series of discrete, stochastic mRNA production events. This transcriptional time series shows intermittent, bursty behavior. One consequence of this temporal intricacy is that gene expression can be tuned by varying different features of the time series. Here we quantify copy-number statistics of mRNA from 20 Escherichia coli promoters using single-molecule fluorescence in situ hybridization in order to characterize the general properties of these transcriptional time series. We find that the degree of burstiness is correlated with gene expression level but is largely independent of other parameters of gene regulation. The observed behavior can be explained by the underlying variation in the duration of bursting events. Using Shannon's mutual information function, we estimate the mutual information transmitted between an outside stimulus, such as the extracellular concentration of inducer molecules, and intracellular levels of mRNA. This suggests that the outside stimulus transmits information reflected in the properties of transcriptional time series.
Figures

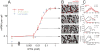

Comment in
-
Transcription: getting close to the action.Nat Rev Genet. 2011 Jun;12(6):382. doi: 10.1038/nrg3008. Epub 2011 May 17. Nat Rev Genet. 2011. PMID: 21577224 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

