Saturation of the human phenome
- PMID: 21532833
- PMCID: PMC3048311
- DOI: 10.2174/138920210793175886
Saturation of the human phenome
Abstract
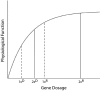
The phenome is the complete set of phenotypes resulting from genetic variation in populations of an organism. Saturation of a phenome implies the identification and phenotypic description of mutations in all genes in an organism, potentially constrained to those encoding proteins. The human genome is believed to contain 20-25,000 protein coding genes, but only a small fraction of these have documented mutant phenotypes, thus the human phenome is far from complete. In model organisms, genetic saturation entails the identification of multiple mutant alleles of a gene or locus, allowing a consistent description of mutational phenotypes for that gene. Saturation of several model organisms has been attempted, usually by targeting annotated coding genes with insertional transposons (Drosophila melanogaster, Mus musculus) or by sequence directed deletion (Saccharomyces cerevisiae) or using libraries of antisense oligonucleotide probes injected directly into animals (Caenorhabditis elegans, Danio rerio). This paper reviews the general state of the human phenome, and discusses theoretical and practical considerations toward a saturation analysis in humans. Throughout, emphasis is placed on high penetrance genetic variation, of the kind typically asociated with monogenic versus complex traits.
Keywords: Human genome; genetics; phenome; saturation mutagenesis..
Figures
References
LinkOut - more resources
Full Text Sources
