Somatic mutagenesis with a Sleeping Beauty transposon system leads to solid tumor formation in zebrafish
- PMID: 21533036
- PMCID: PMC3080878
- DOI: 10.1371/journal.pone.0018826
Somatic mutagenesis with a Sleeping Beauty transposon system leads to solid tumor formation in zebrafish
Abstract
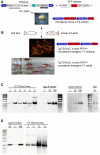
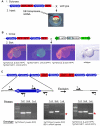
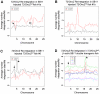
Large-scale sequencing of human cancer genomes and mouse transposon-induced tumors has identified a vast number of genes mutated in different cancers. One of the outstanding challenges in this field is to determine which genes, when mutated, contribute to cellular transformation and tumor progression. To identify new and conserved genes that drive tumorigenesis we have developed a novel cancer model in a distantly related vertebrate species, the zebrafish, Danio rerio. The Sleeping Beauty (SB) T2/Onc transposon system was adapted for somatic mutagenesis in zebrafish. The carp ß-actin promoter was cloned into T2/Onc to create T2/OncZ. Two transgenic zebrafish lines that contain large concatemers of T2/OncZ were isolated by injection of linear DNA into the zebrafish embryo. The T2/OncZ transposons were mobilized throughout the zebrafish genome from the transgene array by injecting SB11 transposase RNA at the 1-cell stage. Alternatively, the T2/OncZ zebrafish were crossed to a transgenic line that constitutively expresses SB11 transposase. T2/OncZ transposon integration sites were cloned by ligation-mediated PCR and sequenced on a Genome Analyzer II. Between 700-6800 unique integration events in individual fish were mapped to the zebrafish genome. The data show that introduction of transposase by transgene expression or RNA injection results in an even distribution of transposon re-integration events across the zebrafish genome. SB11 mRNA injection resulted in neoplasms in 10% of adult fish at ∼10 months of age. T2/OncZ-induced zebrafish tumors contain many mutated genes in common with human and mouse cancer genes. These analyses validate our mutagenesis approach and provide additional support for the involvement of these genes in human cancers. The zebrafish T2/OncZ cancer model will be useful for identifying novel and conserved genetic drivers of human cancers.
Conflict of interest statement
Figures


References
-
- Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability–an evolving hallmark of cancer. Nat Rev Mol Cell Biol. 2010;11:220–228. - PubMed
-
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–627. - PubMed
-
- Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. - PubMed
-
- Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

